Emerging Oncogenic Viruses in Head and Neck Cancers from Romanian Patients
Ramona Gabriela Ursu 1 , Ionut Luchian 2,* , Nicolae Ghetu 3,* , Victor Vlad Costan 4 , Ovidiu Stamatin 4, Octavian Dragos Palade 5 , Costin Damian 1,*, Luminita Smaranda Iancu 1,†and Elena Porumb-Andrese 6
1 Discipline of Microbiology, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Iasi, Romania; ramona.ursu@umfiasi.ro (R.G.U.); luminita.iancu@umfiasi.ro (L.S.I.)
2 Department of Periodontology, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Iasi, Romania
3 Department of Plastic Surgery, Regional Oncology Institute, 700483 Ias¸i, Romania
4 Discipline of Oro-Maxillo-Facial Surgery, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Iasi, Romania; victor.costan@umfiasi.ro (V.V.C.); ovidiu.stamatin@umfiasi.ro (O.S.)
5 Department of ENT, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Iasi, Romania;
octavian.palade@umfiasi.ro
6 Department of Dermatology, “Sf. Spiridon” Clinical Emergency County Hospital, 700111 Iasi, Romania; elena.andrese@umfiasi.ro
* Correspondence: ionut.luchian@umfiasi.ro (I.L.); ghetu.nicolae@umfiasi.ro (N.G.);
costin.v.damian@students.umfiasi.ro (C.D.)
† Author with equal contribution as the first author.
Abstract: (1) Background: Head and neck squamous cell carcinomas (HNSCCs) are some of the most frequent malignancies globally. Oncogenic viruses MCPyV, EBV and HPVs are recognized to be related to HNSCCs and skin cancers. There are no data from Romania regarding the involvement of herpes viruses and polyomaviruses in these types of cancer. We aim to evaluate the association of oncogenic viruses from Papillomaviridae, Herpesviridae, and Polyomaviridae families in HNSCCs and skin cancers. (2) Methods: A total of 26 fresh tumors (6/26 women) were tested for 67 viral agents using a multiplex PCR genotyping assay. (3) Results: A total of 23/26 (88.5%) samples were positive for one or more viruses. All the tested tumors were negative for any HPV (alpha or beta types). In total, we detected as positive samples: 16 (61.63%) EBV1, 12 (46.15%) HHV7, 8 (30.76%) MCV, 6 (23.07%) CMV and HHV6, 2 (7.69%) HHV8, 1 (3.8%) HPyV6 and EBV2. (4) Conclusions: We detected HPV-negative cases that are HPyV and HHV positive. In these fractions of HPV-negative HNSCCs cases, other oncogenic viruses may be involved, such as EBV1, MCV or CMV. Additional research is required for clarifying the natural history of these viruses in HNSCCs, as virus detection would have adecisive impact on diagnostic and decisional algorithms.
Keywords: oncogenic virus; criteria for carcinogenicity; screening; clinical utility
1.Introduction
Cancer cases are one of the most important cause of death in the EU, with 3.5 million new cases diagnosed each year, leading to 1.3 million deaths. The current estimation is that, up to 2035, incidence will increase by 24%, up to 40% for cancer cases which could be prevented [1]. Romania has a high burden of cancer. The estimated age-standardized incidence rates in 2020, for hypopharynx, larynx, lip, oral cavity, both sexes, all ages in Romania were 13.0 ASR, being the third highest in Europe, and for non-melanoma skin cancer was 7.3 ASR (World), in 25th place in Europe [2–4]. These updated data, which are available on the International Agency for Research on Cancer (IARC)—Global Cancer Registry underline the importance of these two categories of cancers in our country.
The carcinogenetic role of some viruses has been well stated by IARC monographs: Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) have a well-established role in liver carcinogenesis, Epstein–Barr Virus (EBV) is known to be responsible for Burkitt lymphoma in parallel with some co-factors, high risk types of HPV (mainly 16, 18, 31, 33, 45, 52) are associated with cervical cancer. Additionally, HPV16 was recognized for having an important role in the carcinogenicity of oropharyngeal cancers [5]. One virus family with oncogenic members that could be causative for the development of tumors is the polyomavirus (PyV) family. The number of known human PyV species has increased rapidly, from two species (JCPyV and BKPyV) detected in 1971, to 13–14 speciesreported since 2007. Most of these viruses were isolated from nasal samples, skin, serum, or stool samples. Despite their recent discovery, many human PyVs are common in the population, and several, including MCPyV, and human PyV 6 and 7 are frequently found in the skin [6]. Polyomaviruses are also analyzed in association with some cancers: Merkel cell polyomavirus (MCV) was first identified in Merkel cell carcinoma,and the primary site of productive BK polyomavirus replication is the kidney [6].
The involvement of oncogenic viruses in HNSCCs and skin cancers has been evaluated by many authors. A research team from Turku, Finland, detected by qPCR viral DNA of SV40, JCV and BKV from HNSCC samples. The JCV was detected in the highest proportion (37%), proving that JCV could be a cofactor for HNSCC [7].
Chen AA et al. tested salivary gland tumors and detected a high prevalence of beta human papillomavirus DNA in malignant tumors. In contrast, polyomavirus DNA was present in all categories of tested samples: cancerous and non-cancerous tissues samples. These viruses are thought to be involved in the carcinogenesis of these tumors, which could eventually be proven by further mechanistic studies [8]. By comparison, a research team from Stockholm did not find any association between salivary gland tumorsand neither HPVs, nor polyomaviruses [9,10].
Regarding Herpesviruses’ involvement in HNSCCs, many studies are referring to EBV and its association with nasopharyngeal cancers, and even more so, some authors are analyzing the possibility of developing a screening test for people at risk, which would detect EBV in liquid biopsies [11].
As for HPV involvement in HNSCC, the scientific literature is abundant, and the prevalence is different for each continent, and is associated with known risk factors (smoking, alcohol). Carlander AF et al. observed a considerable difference regarding HPV prevalence in HNSCC globally, with a general tendency of increasing HPV+ oropharyngeal cancers [12].
Klufah F. et al. detected polyomaviruses in skin cancers, but the authors recognized the necessity of another carcinogenic marker to support the etiologic role of the viruses [13]. Zhao Y. et al. analyzed the association between ultraviolet radiation (UVR) and cutaneous viral infections: beta HPV types andpolyomaviruses. This association seems to favor skin cancer development [14].
The above-mentioned literature, published by known researchers in the field, presents the possible association between these oncogenic viruses and the HNSCCs and skin cancers. We have also tested, so far, different tumor samples for oncogenic viruses, e.g., HPV only [9,15,16], for polyomaviruses only [10], for HBV only [17], but we have never tested, until now, tumor samples for viruses belonging to the three different viral families simultaneously.
The results of this study will help to understand the mechanism of these cancers, and if the viral etiology could be confirmed, at least as a co-factor. As a consequence, our results can contribute to the development of new therapeutic strategies. A tumor induced by a virus could be efficiently prevented by vaccination or treated by oncolytic viral therapy and/or by targeted therapy, in parallel with preventive measures for other co-factors.
Aim: to evaluate the association between DNA of oncogenic viruses from Papillo- maviridae,Herpesviridae, and Polyomaviridae families in different HNSCCs and skin cancers from Romanian patients,using a sensitive molecular assay.
2.Materials and Methods
2.1. Samples
A total of 26 fresh tumor samples (6/26 originating from women) were collected during interventions from surgical departments Iasi, Romania (Oral and Maxillofacial surgery, ENT surgery, Plastic surgery) and then confirmed by histopathology analysis. The median age (range) of the tested patients was 60 (36–78) years. Histological analyses revealed two particular tumors: a recurrent maxillary ameloblastoma of the genian region(52 years old patient), and a solid, ulcerated basal cell carcinoma of the upper lip (another 52 years old patient). All the other tumors were diagnosed as squamous cell carcinoma with keratinization, moderately differentiated. A total of 6/26 (23.07%) of patients were registered as presenting metastases. An amount of 5/26 (19.23%) of patients lived in an urban area. All the patients graduated from gymnasium or professional school. A total of 11/26 (42.3%) declared smoking and 10/26 (38.4%) declared chronic alcohol consumption.None of the tested patients had any other neoplasia or any organ transplant previously. One single patient,smoker, chronic alcohol consumer, with lower lip carcinoma, declared daily exposure to UV light, given his work conditions. We did not find any data in the hospital registries regarding chemo or radiotherapy treatments. The tumor samples were shipped in Eppendorf tubes with RNAlater solution to the Infections and Cancer Biology Group, Section of Infections, IARC, Lyon, France. The study was approved by the Ethical Commission of our university (reference number 7150). All the samples were anonymously tested, and all thepatients signed an informed consent form.
2.2. DNA Extraction
For each sample, sterile scalpels were used to cut the tumors into small pieces. In short, the steps for DNA purification were: incubation of tumor tissue in digestion buffer overnight, at 37 ◦C, then incubation for 10 min at 95 ◦C, centrifugation for 2 min at 15,700× g, chilling on ice, to inactivate the proteinase K. New sterile Eppendorf tubes were used for the final aqueous phase. No contamination was detected throughout the study.
2.3. Detection of Viral DNA
A total of 67 infectious agents were detected by a type-specific multiplex genotyping (TS-MPG) assay, which combines multiplex polymerase chain reaction (PCR) and bead- based assay using the Luminex®100/200™ system (Luminex Corp., Austin, TX, USA), as described previously [18–20]. Multiplex type-specific PCR uses specific primers for the detection of 19 probable/high-risk alpha-HPV types (HPV 16, 18,26, 31, 33, 35, 39, 45, 51,52, 53, 56, 58, 59, 66, 68a, 68b, 70, 73, and 82), 2 low-risk alpha-HPV types (HPV 6, 11), 25 genus-beta HPV types (HPV 5, 8, 9, 12, 14, 15, 17, 19, 20, 21, 22, 23, 24, 25, 36, 37, 38, 47, 49, 75, 76, 80, 92, 93, and 96) from the Papillomaviridae family, 12 polyomaviruses (JCV-John Cunningham virus, BKV, KIV-Karolinska Institute Virus, WUV-Washington University Virus, MCV-Merkel Cell Polyomavirus, HPyV6, HPyV7, TSV-Trichodysplasia Spinulosa Polyomavirus, HPyV9, HPyV10, HPyV12, SV40-Simian Virus 40) from the Polyomaviridae family, and 9 herpesviruses: HSV1 (HHV1), HSV2 (HHV2),varicella zoster virus (HHV3), EBV (HHV4) with EBV1 and EBV2 types, CMV (HHV5), human herpesvirus 6 (HHV6), human herpesvirus 7 (HHV7), HHV8 (KSHV or Kaposi’s sarcoma-associated herpesvirus) from theHerpesviridae family [21,22]. Beta-globin primers were used like a positive control to assess the DNA quality.
2.4. Statistical Analysis
All the statistical analyses were performed using SPSS version 24.0 software (IBM Corp., Armonk, NY, USA). Qualitative significance tests, such as Chi-square tests, were used for comparing the distributions off requencies.
3. Results
A total of 23/26 (88.5%) patients were positive for one or more viruses. In total, we detected as positive samples: 16 (61.63%) EBV1, 12 (46.15%) HHV7, 8 (30.76%) MCV and HHV6, 6 (23.07%) CMV, 2 (7.69%) HHV8, 1 (3.8%) HPyV6 and EBV2 (Figure 1). We detected tumors with unique viral infections (six), with double infections (four), with triple viral infections (eight) and four tumors were positive for DNA of four of these viruses.
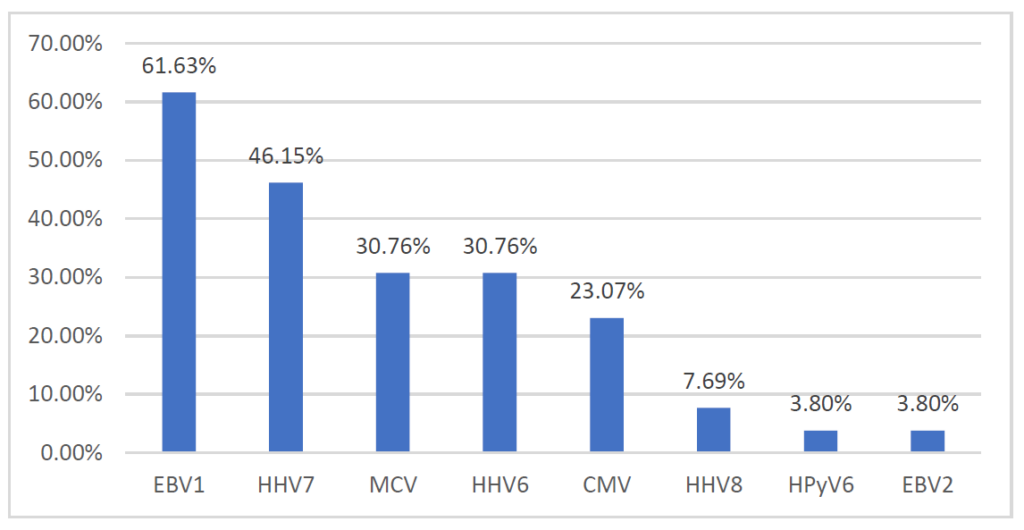
Figure 1. The prevalence of tested viruses in tumor samples.
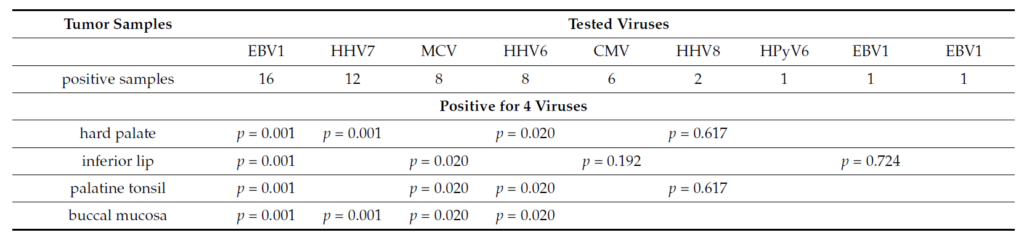
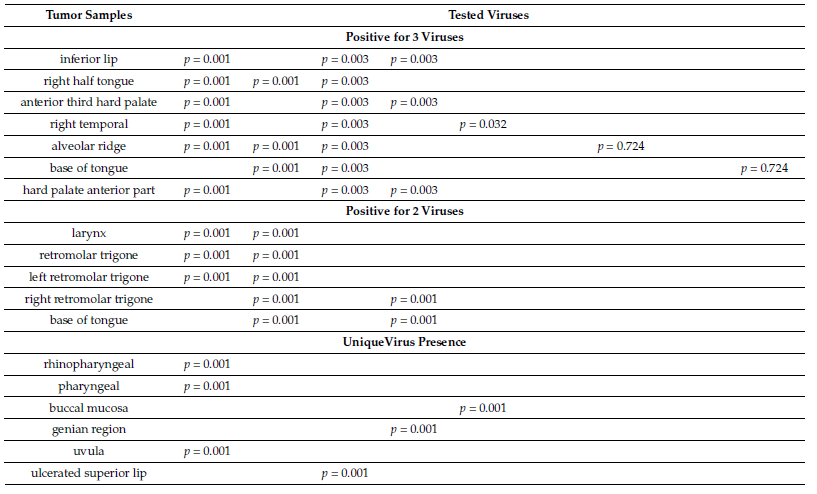
The tumor samples which were detected positive for four viruses were from hard palate (EBV1, HHV6, HHV7, HHV8), inferior lip (MCV, EBV1, CMV, HHV7), palatine tonsil (MCV, EBV1, HHV6, HHV8), and right buccal mucosa (MCV, EBV1, HHV6, HHV7). The triple virus presence was detected in the next cancers: inferior lip (MCV, EBV1, CMV), right half tongue (EBV1, CMV, HHV7), anterior third hard palate (MCV, EBV1, HHV6), right temporal (EBV1, CMV, HHV7), alveolar ridge (HPyV6, EBV1, HHV7), base of tongue(MCV, EBV2, HHV7), and hard palate anterior part (MCV, EBV1, HHV6). The double virus presence was detected in the next tumors: larynx (EBV1, HHV7), retromolar trigone (EBV1, HHV7), left retromolar trigone (EBV1, HHV7), right retromolar trigone (HHV6, HHV7), and base of tongue (HHV6, HHV7). The single virus presence was seen in the following cancers: rhinopharyngeal (EBV1), pharyngeal (EBV1), buccal mucosa with skin extension (CMV), genian region (HHV6), uvula (EBV1), ulcerated superior lip (MCV). The highest detected viral load was detected for HHV8, in both cases (hard palate and glottis cancers). We detected significant association between the tested tumors and EBV1, HHV7, MCV, HHV6, CMV for multiple viraldetection in the tissue samples (p < 0.05) (Table 1).
Table 1. Correlations between tumor samples and the number of positive viruses.
4.Discussions
In this pilot study, we tested HNSCC and skin tumor samples using a very sensitive assay for detecting very small amounts of DNA in a multiplex design. The method was optimized to detect 67 etiologic agents simultaneously from Papillomaviridae, Herpesviridae and Polyomaviridae families. The assay has the advantage of testing many viral etiologies in a short period of time, by a completely automatized method,which removes any source of possible errors. The results of the testing were validated by the quality controls used. To compare our experimental results with other research data, we reviewed the recent scientific literature regarding these viruses’ involvement in the carcinogenesis of HNSCCs. Herpes viruses are known to have an oncogenic role [5]. The highest prevalence of the viruses we tested was for EBV1 (61.63%) and EBV2was detected in fewer samples (3.8%). Therefore, we are mentioning Lee LA et al., who detected using RealTime PCR a 48% EBV prevalence in Primary Laryngeal Squamous Cell Carcinoma, from Formalin-fixedparaffin- embedded (FFPE) tissue sections [23]. Deng Z et al. analyzed the prevalence and prognostic values of EBV infections in Japanese patients with different types of HNSCC from fresh frozen samples, using PCR.The authors detected DNA/EBV in 69.9% HNC cases, from which 73.3% was EBV1 and 18.5% EBV2.Even though EBV was highly prevalent in cancers of the oropharynx and nasopharynx, EBV could not beused as a prognostic marker for the studied cohort [24]. Both EBV types were detected in another study which analyzed EBV in FFPE HNSCC tissue samples by PCR and confirmed by nucleotide sequencing. The authors detected a 12.8% EBV prevalence with 90% EBV1 and 10% EBV2 [25]. Zhou X et al. used EBV markers to develop a risk score for nasopharyngeal carcinoma which opens new opportunities for personalized risk prediction and for an early diagnosis of nasopharyngeal carcinoma [26]. Another team of researchers used the viral load of DNA/EBV detected in 75% of their tested nasopharyngeal carcinoma samples as a prognosis marker for a better disease-free survival [27]. A very recent case (September 2021) was published by a team of researchers from Canada: the authors described a 78-year-old Chinese male diagnosed with a primary EBV-positive, HPV-negative nasopharyngeal carcinoma, in the left tonsil, as being the first case of primary tonsil EBV-positive nasopharyngeal carcinoma described in the literature [28]. Our results for EBV detection are comparable with the data published by the above-mentioned studies.
HHV8 was detected in all 11 head and neck Kaposi Sarcoma cases, analyzed by immunostaining [29]. HHV8 was also detected, by other authors, in oropharyngeal cancers [30] by immunohistochemistry, and in rare cases of nasopharyngeal cancer, by histology and immunohistochemical markers [31]. Our two cases detected positive for HHV8 are in line with the above cited papers.
HHV6 and HHV7 were tested by PCR, and an oral shedding of HHV6 and HHV7 was observed, during all the radiotherapy period for the tested patients [32]. HHV6 was found in 16.7% of adult-onset recurrent respiratory papillomatosis biopsies, by RT PCR [33]. Our results show that we detected these viruses in a higher proportion than the ones published by other authors: 46.15% HHV7, and 30.76% HHV6.
Kiprian D et al. quantified the DNA/CMV in plasma of patients with HNSCC under radiotherapy and chemotherapy, and for their study patients they found that plasma positivity was associated with a higher risk of death [34]. Radunovic M et al. analyzed the CMV antigen in salivary glands tumors (70.6%), and they found that CMV might play an important role in the development of these kinds of tumors [35]. Gangemi A.C. et al. recently published a case of a HIV-negative patient with oropharyngeal cancer in remission, which developed a buccal ulcer positive for CMV. The authors conclude that opportunistic infections with CMV can affect the quality of life and mortality [36]. The above papers support our 23.07% CMV prevalence detected and its possible role as a carcinogenic etiologic agent.
Regarding MCV we detected a 30.76% MCV presence in the tested tumors. The analyzed studies are different regarding their findings. A research team from the Nether- lands [37] tested polyomaviruses in patients with HNSCC non-smokers and non-drinkers using tissue microarrays and immunohistochemistry, and MCV was not detected in any of the cases. Another research team from Chile [38] assayed HNSCC using PCR and they detected MCV in 12.5% of tested cases. Dickinson A et al., from Finland, evaluated oropharyngeal tumors by Real-Time PCR and detected MCV in the tissue for one single person [39]. Besides the simple presence of MCV in the tumor tissues, other authors evaluated the role of this virus as a co-carcinogenic factor. Herberhold S. et al., from Germany, tested and detected MCV in 10.0% of tonsillar squamous cell carcinomas, but the lowviral load detected by real-time PCR did not support the role of polyomaviruses in tonsillar carcinogenesis [40].Saláková M et al. detected MCV by qPCR in 10.2% of the tonsillar carcinomas tested, with the same low viral load as the previous authors [41]. Carpén T. et al. analyzed all the polyomaviruses in oropharyngeal squamouscell carcinoma by PCR and they detected DNA of BKV, JCV and SV40, but the authors concluded that thepresence of these viruses has no impact on survival [42].
Regarding HPV, it is well recognized by IARC that HPV 16 is involved in oropharyngeal carcinogenesis [5]. The fact that we did not detect any HPV-positive cases in this study was not a surprise for us, as we have already published two studies in which we did not find any HPV-positive case from 20 base of tongue cancers and tonsil cancers [43], and in a second larger study, we detected just 12.2%/189 cases HPV DNA positive, from which only 1.1% were HPV-driven (positive for HPV DNA and RNA) [44]. In a recent review published by Carlander AF et al. [12], the authors mention the considerable variation in HPV prevalence: 10% in Spain [45], and 70% in Sweden [46]. The authors reviewed the studies which used double HPV/DNA and p16 algorithms testing for oropharyngeal cancers. The same significant variation of HPV HNSCC positive cases worldwide was detected previously by two other research teams, with the suggestion that HPV 16 is very important mainly for oropharyngeal carcinogenesis [47,48].
The beta HPV types were detected by a research team from the USA, using next-generation sequencing(NGS) from patients with oral cavity and oropharyngeal cancer cases [49]. Galati L et al. detected beta HPV types using Luminex-based assays and NGS in conjunctivalsquamous cell cancers [50].
The scientific literature cited above presented the results of studies realized for a single oncogenic virus or viruses from two viral families, using different diagnosis as- says, including PCR, Real Time PCR, NGS,and immunohistochemistry. This brief review proves the difficult elements in the research of this issue, focusing on the possible involvement of viruses in the head and neck carcinogenetic mechanism, and the limitations of current approaches.
We detected an increased prevalence (88.5%) of the known oncogenic viruses, other than HPVs. These results are in line with others of our studies, in which we detected a low HPV prevalence in HNSCCs [43,44], ona more representative number of samples. For this category of patients, we could think of other risk cofactors,such as smoking (42.3%) and chronic alcohol consumption (38,4%), as the patients declared. The high prevalence of the other detected viruses (e.g., 61.63% EBV1, 30.76% MCV and 23.07% CMV) in these tumorsamples could help to understand the mechanism of these cancers, and if this viral etiology could be confirmed, at least as co-factor.
We need to mention that the simple presence of DNA of one known oncogenic virus is not proving the etiological role of that virus in a specific tumor tissue. We would need to test some other markers to support this etiologic role, like in other previous studies of ours (e.g., viral RNA testing, p16/p53 by immunohistochemistry assays) [43,44].
Beside the presence and persistence of viral DNA in tumor biopsies, the following characteristics have been proposed to define human viruses that cause cancer through direct or indirect carcinogenesis: growth-promoting activity of viral genes in model systems; dependence of a malignant phenotype on continuous viral oncogene expression or modification of host genes; and epidemiological evidence that a virus infection represents a major risk for the development of cancer [51].
The clinical utility of detecting oncogenic viruses in tumors relates to specific prevention, as for some of the viruses there are already available vaccines (bi-, tetra- and nonavalent against HR HPV types), while for some other viral induced tumors therapeutic vaccines could be developed [52].
For HPV viruses, the screening algorithm for cervical cancer is already well established— Pap test and high-risk HPV type testing [16], but less so for HNSCCs. Additionally, for some tumors which are viral-induced, targeted therapy with FGFR and PIK3 inhibitors is available [53].
Patients with HPV-positive TSCC and BOTSCC have a much better prognosis than those with corresponding HPV-negative cancers, and the incidence of HPV-positive cancers has been increasing epidemically in the past decades. Tailoring therapy for this growing group of patients is crucial, and efforts in finding markers useful for tapering and targeting therapy are on the way and will hopefully be of benefit forsome patients [54].
A limitation of the study is that we could not include a more significant number of tumor samples, due to the fact that this research was part of a pilot study, in which this complex assay was used for the first time. The results of this research warrant the need to undergo further studies, in order to evaluate the clinical utility of testing tumors for known oncogenic viruses.
Our results could contribute to the development of new therapeutic strategies in the management oftumors which are positive for polyomaviruses and herpesviruses.
5.Conclusions
The high prevalence of viral infections in the tested patients (EBV1, HHV7, HHV6, CMV, MCV), other than HPVs, suggests an association between the presence of viral DNA of these oncogenic viruses and HNSCCs. Additional research is required to clarify the natural history of these viruses in HNSCCs, as virus detection would have a decisive impact on diagnostic and decisional algorithms.
Author Contributions: Conceptualization: R.G.U.; methodology, R.G.U. and E.P.-A.; resources, I.L., N.G., V.V.C., O.S.and O.D.P.; writing—R.G.U. original draft preparation, R.G.U. and C.D.; writing—
R.G.U. and C.D.; review and editing, R.G.U., C.D. and L.S.I. All authors have read and agreed to the published version of the manuscript.
Funding: The study was undertaken by R.G. Ursu while hosted as Visiting Scientist by the Inter- national Agency for Research on Cancer, Lyon, France. Scientific research funded by University of Medicine and Pharmacy “Grigore TPopa”, Iasi, research contract 6983 from 21 April 2020.
Institutional Review Board Statement: The study was conducted according to the guidelines of the Declaration of Helsinki and approved by Ethics Committee of “Grigore T. Popa” University of Medicine and Pharmacy(7150/10.02.2015).
Informed Consent Statement: Informed consent was obtained from all subjects involved in the study.
Data Availability Statement: Data is contained within the article.
Acknowledgments: We are grateful to Massimo Tommasino and Tarik Gheit who gave permission to Ursu to test these tumor samples at IARC, during her internship program as Visiting Scientist at Infections and Cancer Biology Group, International Agency for Research on Cancer, Lyon, France. Part of data from this article have been presented at IARC 50th Anniversary Conference “Global Cancer: Occurrence, Causes, and Avenues to Prevention”, Centre de Congrès, Lyon, France, 7–10 June 2016.
Conflicts of Interest: The authors declare no conflict of interest.
References
- Europe Beating Cancer Plan. Joint Research Center. 2020. Available online: https://ec.europa.eu/info/law/better-regulation/ have-your-say/initiatives/12154-Europe%E2%80%99s-Beating-Cancer-Plan_en (accessed on 21 August 2021).
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. 2020. Available online: https://gco.iarc.fr/today (accessed on 21 August 2021).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN estimates ofincidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [CrossRef]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence andmortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2018, 144, 1941–1953. [CrossRef] [PubMed]
- International Agency for Research on Cancer. A Review of Human Carcinogens: Biological Agents. IARC Monographs of the Evaluation ofCarcinogenic Risks to Humans; IARC: Lyon, France, 2012.
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Malaria and Some Polyomaviruses (SV40, BK, JC, and Merkel CellViruses). IARC Monographs on the Evaluation of Carcinogenic Risks to Humans; IARC: Lyon, France, 2014.
- Poluschkin, L.; Rautava, J.; Turunen, A.; Wang, Y.; Hedman, K.; Syrjänen, K.; Grenman, R.; Syrjänen, S. Polyomaviruses detectable in head andneck carcinomas. Oncotarget 2018, 9, 22642–22652. [CrossRef] [PubMed]
- Chen, A.A.; Gheit, T.; Stellin, M.; Lupato, V.; Spinato, G.; Fuson, R.; Menegaldo, A.; Mckay-Chopin, S.; Cin, E.D.; Tirelli, G. Oncogenic DNAviruses found in salivary gland tumors. Oral Oncol. 2017, 75, 106–110. [CrossRef] [PubMed]
- Haeggblom, L.; Ursu, R.G.; Mirzaie, L.; Attoff, T.; Gahm, C.; Nordenvall, L.H.; Nasman, A. No evidence for human papillomavirus having acausal role in salivary gland tumors. Diagn. Pathol. 2018, 13, 44. [CrossRef]
- Ramqvist, T.; Ursu, R.G.; Haeggblom, L.; Mirzaie, L.; Gahm, C.; Hammarstedt-Nordenvall, L.; Dalianis, T.; Nasman, A. Human polyomaviruses arenot frequently present in cancer of the salivary glands. Anticancer. Res. 2018, 38, 2871–2874. [CrossRef]
- Tan, R.; Phua, S.K.A.; Soong, Y.L.; Oon, L.L.E.; Chan, K.S.; Lucky, S.S.; Mong, J.; Tan, M.H.; Lim, C.M. Clinical utility of Epstein-Barr virus DNA and other liquid biopsy markers in nasopharyngeal carcinoma. Cancer Commun. 2020, 40, 564–585. [CrossRef]
- Carlander, A.F.; Jakobsen, K.K.; Bendtsen, S.K.; Garset-Zamani, M.; Lynggaard, C.D.; Jensen, J.S.; Grønhøj, C.; Buchwald, C. A contemporary systematic review on repartition of HPV-positivity in oropharyngeal cancer worldwide. Viruses 2021, 13, 1326. [CrossRef]
- Klufah, F.; Mobaraki, G.; Liu, D.; Alharbi, R.A.; Kurz, A.K.; Speel, E.J.M.; Winnepenninckx, V.; Zur Hausen, A. Emerging role of humanpolyomaviruses 6 and 7 in Human Cancers. Infect. Agents Cancer 2021, 16, 1–12. [CrossRef]
- Zhao, Y.; Amorrortu, R.P.; Fenske, N.A.; Cherpelis, B.; Messina, J.L.; Sondak, V.K.; Giuliano, A.R.; Schell, M.J.; Waterboer, T.; Pawlita, M.Cutaneous viral infections associated with ultraviolet radiation exposure. Int. J. Cancer 2021, 148, 448–458. [CrossRef]
- Ursu, R.G.; Ivanov, I.; Popescu, E.; Costan, V.; Stamatin, O.; Ghetu, N.; Palade, D.; MIRTU, C.; Andrese, E.; Danciu, M. Human papilloma virusgenotyping in fresh head and neck tumors-our first experience. Med. Surg. J. 2015, 119, 676–680.
- Ursu, R.G.; Onofriescu, M.; Luca, A.; Prisecariu, L.J.; Salceanu, S.O.; Nemescu, D.; Iancu, L.S. The need for cervical cancer control in HIV-positiveand HIV-negative women from romania by primary prevention and by early detection using clinically validated HPV/DNA tests. PLoS ONE 2015, 10,e0132271. [CrossRef]
- Popovici, O.; Ursu, R.G.; Azoicai, D.; Iancu, L.S. HBV genotypes circulation in pregnant women in Romania: A pilot study. Rev. Romana Med. Lab.2020, 28, 91–98. [CrossRef]
- Schmitt, M.; Dondog, B.; Waterboer, T.; Pawlita, M.; Tommasino, M.; Gheit, T. Abundance of multiple high-risk human papillomavirus (HPV) infections found in cervical cells analyzed by use of an ultrasensitive HPV genotyping assay. J. Clin. Microbiol. 2010, 48, 143–149. [CrossRef][PubMed]
- Gheit, T.; Billoud, G.; de Koning, M.N.; Gemignani, F.; Forslund, O.; Sylla, B.S.; Vaccarella, S.; Franceschi, S.; Landi, S.; Quint, W.G. Development of a sensitive and specific multiplex PCR method combined with DNA microarray primer extension to detect Betapapillomavirustypes. J. Clin. Microbiol. 2007, 45, 2537–2544. [CrossRef] [PubMed]
- Gheit, T.; Landi, S.; Gemignani, F.; Snijders, P.J.; Vaccarella, S.; Franceschi, S.; Canzian, F.; Tommasino, M. Development of a sensitive and specific assay combining multiplex PCR and DNA microarray primer extension to detect high-risk mucosal human papillomavirus types. J. Clin.Microbiol. 2006, 44, 2025–2031. [CrossRef] [PubMed]
- Schmitt, M.; Bravo, I.G.; Snijders, P.J.; Gissmann, L.; Pawlita, M.; Waterboer, T. Bead-based multiplex genotyping of human Papillomaviruses. J.Clin. Microbiol. 2006, 44, 504–512. [CrossRef]
- Corbex, M.; Bouzbid, S.; Traverse-Glehen, A.; Aouras, H.; McKay-Chopin, S.; Carreira, C.; Lankar, A.; Tommasino, M.; Gheit, T. Prevalence of papillomaviruses, polyomaviruses, and herpesviruses in triple-negative and inflammatory breast tumors from algeria compared with other types ofbreast cancer tumors. PLoS ONE 2014, 9, e114559. [CrossRef]
- Lee, L.A.; Fang, T.J.; Li, H.Y.; Chuang, H.H.; Kang, C.J.; Chang, K.P.; Liao, C.T.; Chen, T.C.; Huang, C.G.; Yen, T.C. Effects of Epstein-Barr virus infection on the risk and prognosis of primary laryngeal squamous cell carcinoma: A hospital-based case-control study in Taiwan. Cancers2021, 13, 1741. [CrossRef]
- Deng, Z.; Uehara, T.; Maeda, H.; Hasegawa, M.; Matayoshi, S.; Kiyuna, A.; Agena, S.; Pan, X.; Zhang, C.; Yamashita, Y.; et al. Epstein-Barr virusand human papillomavirus infections and genotype distribution in head and neck cancers. PLoS ONE 2014, 9, e113702. [CrossRef]
- Niya, M.H.K.; Tameshkel, F.S.; Keyvani, H.; Esghaei, M.; Panahi, M.; Zamani, F.; Tabibzadeh, A. Epstein-Barr virus molecular epidemiology and variants identification in head and neck squamous cell carcinoma. Eur. J. Cancer Prev. 2020, 29, 523–530. [CrossRef]
- Zhou, X.; Cao, S.M.; Cai, Y.L.; Zhang, X.; Zhang, S.; Feng, G.F.; Chen, Y.; Feng, Q.S.; Chen, Y.; Chang, E.T.; et al. A comprehensive risk score for effective risk stratification and screening of nasopharyngeal carcinoma. Nat. Commun. 2021, 12, 5189. [CrossRef] [PubMed]
- Nilsson, J.S.; Forslund, O.; Andersson, F.C.; Lindstedt, M.; Greiff, L. Intralesional EBV-DNA load as marker of prognosis for nasopharyngealcancer. Sci. Rep. 2019, 9, 15432. [CrossRef] [PubMed]
- Nguyen, S.; Kong, T.; Berthelet, E.; Ng, T.; Prisman, E. A Unique Case of Primary EBV-Positive, HPV-Negative Nasopharyngeal Carcinoma Locatedin the Tonsil. Head Neck Pathol. 2021, 15, 1017–1022. [CrossRef] [PubMed]
- Agaimy, A.; Mueller, S.K.; Harrer, T.; Bauer, S.; Thompson, L.D.R. Head and neck Kaposi sarcoma: Clinicopathological analysis of 11 cases.Head Neck Pathol. 2018, 12, 511–516. [CrossRef] [PubMed]
- Sikora, A.G.; Shnayder, Y.; Yee, H.; DeLacure, M.D. Oropharyngeal kaposi sarcoma in related persons negative for human immunodeficiencyvirus. Ann. Otol. Rhinol. Laryngol. 2008, 117, 172–176. [CrossRef] [PubMed]
- Soon, G.S.T.; Petersson, F.; Thong, M.K.T.; Tan, C.L. Primary nasopharyngeal Kaposi sarcoma as index diagnosis of AIDS in a previously healthyman. Head Neck Pathol. 2018, 13, 664–667. [CrossRef] [PubMed]
- Palmieri, M.; Martins, V.A.O.; Sumita, L.M.; Tozetto-Mendoza, T.R.; Romano, B.B.; Machado, C.M.; Pannuti, C.S.; Brandão, T.B.; Ribeiro, A.C.P.; Corrêa, L.; et al. Oral shedding of human herpesviruses in patients undergoing radiotherapy/chemotherapy treatment for head and neck squamous cellcarcinoma. Clin. Oral Investig. 2017, 21, 2291–2301. [CrossRef]
- Formánek, M.; Formánková, D.; Hurník, P.; Vrtková, A.; Komínek, P. Epstein-Barr virus may contribute to the pathogenesis of adult-onsetrecurrent respiratory papillomatosis: A preliminary study. Clin. Otolaryngol. 2021, 46, 373–379. [CrossRef]
- Kiprian, D.; Czarkowska-Paczek, B.; Wyczalkowska-Tomasik, A.; Paczek, L. Human cytomegalovirus and Epstein-Barr virus infections increase the risk of death in patients with head and neck cancers receiving radiotherapy or radiochemotherapy. Medicine 2018, 97, e13777. [CrossRef]
- Radunovic, M.; Tomanovic, N.; Novakovic, I.; Boricic, I.; Milenkovic, S.; Dimitrijevic, M.; Radojevic-Skodric, S.; Bogdanovic, L.; Basta-Jovanovic, G. Cytomegalovirus induces Interleukin-6 mediated inflammatory response in salivary gland cancer. J. BUON 2016, 21, 1530–1536.
- Gangemi, A.C.; Choi, S.H.; Yin, Z.; Feurdean, M. Cytomegalovirus and herpes simplex virus co-infection in an HIV-negative patient: A case report.Cureus 2021, 13, 13214. [CrossRef]
- Mulder, F.J.; Klufah, F.; Janssen, F.M.E.; Farshadpour, F.; Willems, S.M.; De Bree, R.; Zur Hausen, A.; Van den Hout, M.F.C.M.; Kremer, B.; Speel, E.M. Presence of human papillomavirus and Epstein-Barr virus, but absence of Merkel cell polyomavirus, in head and neck cancer of non-smokers andnon-drinkers. Front. Oncol. 2021, 10, 560434. [CrossRef] [PubMed]
- Muñoz, J.P.; Blanco, R.; Osorio, J.C.; Oliva, C.; Diaz, M.J.; Carrillo-Beltrán, D.; Aguayo, R.; Castillo, A.; Tapia, J.C.; Calaf, G.M.; et al. Merkel cell polyomavirus detected in head and neck carcinomas from Chile. Infect. Agent. Cancer 2020, 15, 4–5. [CrossRef] [PubMed]
- Dickinson, A.; Xu, M.; Silén, S.; Wang, Y.; Fu, Y.; Sadeghi, M.; Toppinen, M.; Carpén, T.; Hedman, K.; Mäkitie, A.; et al. Newly detected DNA viruses in juvenile nasopharyngeal angiofibroma (JNA) and oral and oropharyngeal squamous cell carcinoma (OSCC/OPSCC). Eur. Arch.Otorhinolaryngol. 2019, 276, 613–617. [CrossRef] [PubMed]
- Herberhold, S.; Hellmich, M.; Panning, M.; Bartok, E.; Silling, S.; Akgül, B.; Wieland, U. Human polyomavirus and human papillomavirus prevalence and viral load in non-malignant tonsillar tissue and tonsillar carcinoma. Med. Microbiol. Immunol. 2017, 206, 93–103. [CrossRef]
- Saláková, M.; Košlabová, E.; Vojteˇchová, Z.; Tachezy, R.; Šroller, V. Detection of human polyomaviruses MCPyV, HPyV6, and HPyV7 inmalignant and non-malignant tonsillar tissues. J. Med. Virol. 2016, 88, 695–702. [CrossRef]
- Carpén, T.; Syrjänen, S.; Jouhi, L.; Randen-Brady, R.; Haglund, C.; Mäkitie, A.; Mattila, P.S.; Hagström, J. Epstein-Barr virus (EBV) andpolyomaviruses are detectable in oropharyngeal cancer and EBV may have prognostic impact. Cancer Immunol. Immunother. 2020, 69, 1615–1626.[CrossRef] [PubMed]
- Bersani, C.; Haeggblom, L.; Ursu, R.G.; Giusca, S.E.; Marklund, L.; Ramqvist, T.; Näsman, A.; Dalianis, T. Overexpression of FGFR3 in HPV-positive tonsillar and base of tongue cancer is correlated to outcome. Anticancer Res. 2018, 38, 4683–4690. [CrossRef]
- Ursu, R.G.; Danciu, M.; Spiridon, I.A.; Ridder, R.; Rehm, S.; Maffini, F.; McKay-Chopin, S.; Carreire, C.; Lucas, E.; Costan, V.V.; et al. Role ofmucosal high-risk human papillomavirus types in head and neck cancers in Romania. PLoS ONE 2018, 13, e0199663. [CrossRef]
- Mena, M.; Frias-Gomez, J.; Taberna, M.; Quirós, B.; Marquez, S.; Clavero, O.; Baena, A.; Lloveras, B.; Alejo, M.; León, X.; et al. Epidemiology of human papillomavirus-related oropharyngeal cancer in a classically low-burden region of southern Europe. Sci. Rep. 2020, 10, 13219. [CrossRef][PubMed]
- Haeggblom, L.; Attoff, T.; Yu, J.; Holzhauser, S.; Vlastos, A.; Mirzae, L.; Ährlund-Richter, A.; Munck-Wikland, E.; Marklund, L.; Hammarstedt-Nordenvall, L.; et al. Changes in incidence and prevalence of human papillomavirus in tonsillar and base of tongue cancer during 2000–2016 in theStockholm region and Sweden. Head Neck 2019, 41, 1583–1590. [CrossRef] [PubMed]
- Anantharaman, D.; Abedi-Ardekani, B.; Beachler, D.C.; Gheit, T.; Olshan, A.F.; Wisniewski, K.; Wunsch-Filho, V.; Toporcov, T.N.; Tajara, E.H.;Levi, J.E.; et al. Geographic heterogeneity in the prevalence of human papillomavirus in head and neck cancer. Int. J. Cancer 2017, 140, 1968–1975.[CrossRef] [PubMed]
- Castellsagué, X.; Alemany, L.; Quer, M.; Halec, G.; Quirós, B.; Tous, S.; Clavero, O.; Alòs, L.; Biegner, T.; Szafarowski, T.; et al. International HPV in Head and Neck Cancer Study Group. HPV involvement in head and neck cancers: Comprehensive assessment of biomarkers in 3680 patients.J. Natl. Cancer Inst. 2016, 108, djv403. [CrossRef]
- Dang, J.; Bruce, G.A.; Zhang, Q.; Kiviat, N.B. Identification and characterization of novel human papillomaviruses in oral rinse samples from oralcavity and oropharyngeal cancer patients. J. Oral Biosci. 2019, 61, 190–194. [CrossRef] [PubMed]
- Galati, L.; Combes, J.D.; Gupta, P.; Sen, R.; Robitaille, A.; Brancaccio, R.N.; Atsou, K.; Cuenin, C.; McKay-Chopin, S.; Tornesello, M.L.; et al. Detection of a large spectrum of viral infections in conjunctival premalignant and malignant lesions. Int. J. Cancer 2020, 147, 2862–2870. [CrossRef]
- Zapatka, M.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sültmann, H.; Moch, H.; Cooper, C.S. The landscape of viralassociations in human cancers. Nat. Genet. 2020, 52, 320–330. [CrossRef]
- Tabachnick-Cherny, S.; Pulliam, T.; Church, C.; Koelle, D.M.; Nghiem, P. Polyomavirus-driven Merkel cell carcinoma: Prospects fortherapeutic vaccine development. Mol. Carcinog. 2020, 59, 807–821. [CrossRef]
- Holzhauser, S.; Wild, N.; Zupancic, M.; Ursu, R.G.; Bersani, C.; Näsman, A.; Kostopoulou, O.N.; Dalianis, T. Targeted therapy with PI3K and FGFR inhibitors on human papillomavirus positive and negative tonsillar and base of tongue cancer lines with and without corresponding mutations.Front. Oncol. 2021, 11, 640490. [CrossRef]
- Näsman, A.; Du, J.; Dalianis, T. A global epidemic increase of an HPV-induced tonsil and tongue base cancer—Potential benefit from a pan-genderuse of HPV vaccine. J. Intern. Med. 2019, 287, 134–152. [CrossRef]