Understanding the Pattern of Oropharyngeal Cancers from North-East Romanian Patients
Ramona Gabriela Ursu 1, Simona Eliza Giusca 2, Irene Alexandra Spiridon 2, Bianca Manole 2,Mihai Danciu 2, Victor Vlad Costan 3,†, Dragos Octavian Palade 4, Nicolae Ghetu 5, Paula Toader 6,Mădălina Alexandra Vlad 1,Costin Damian 1, Elena Porumb-Andrese 7,*, Ionut Luchian 8,* Luminita Smaranda Iancu 1
1.Department of Preventive Medicine and Interdisciplinarity (IX)—Microbiology, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Ias¸i, Romania;ramona.ursu@umfiasi.ro (R.G.U.);madalina-alexandra.gagauta-vlad@d.umfiasi.ro (M.A.V.); costin.damian@d.umfiasi.ro (C.D.); luminita.iancu@umfiasi.ro (L.S.I.)
2.Department of Morphofunctional Sciences I—Pathology, “Grigore T. Popa” University of Medicine andPharmacy, 700115 Ias¸i, Romania; simona-eliza.giusca@umfiasi.ro (S.E.G.); spiridon.irene@umfiasi.ro (I.A.S.); bianca.manole@umfiasi.ro (B.M.);mihai.danciu@umfiasi.ro (M.D.)
3.Discipline of Oro-Maxillo-Facial Surgery, “Grigore T. Popa” University of Medicine and Pharmacy,700115 Ias¸i, Romania; victor.costan@umfiasi.ro
4.Department of ENT, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Iasi, Romania; octavian.palade@umfiasi.ro
5.Department of Plastic Surgery, Regional Oncology Institute, 700483 Ias¸i, Romania; ghetu.nicolae@umfiasi.ro
6.Department of Oral Dermatology, University of Medicine and Pharmacy, 700115 Ias¸i, Romania; mihaela.toader@umfiasi.ro
7.Department of Medical Specialties (III)—Dermatology, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Ias¸i, Romania
8.Department of Periodontology, “Grigore T. Popa” University of Medicine and Pharmacy, 700115 Ias¸i, Romania
*Correspondence: elena.andrese@umfiasi.ro (E.P.-A.); ionut.luchian@umfiasi.ro (I.L.)
† Author with equal contribution as the first author.
Abstract: Background: Human papilloma virus (HPV) is acknowledged as a risk factor for oropha- ryngeal squamous cellular cancers (OPSCC), of which the dominant types are tonsillar (TSCC) and base of tongue cancer (BOTSCC). Objective: To assess the role of HPV in selected OPSCC cases, from Romanian patients by sensitive and complementary molecular assays. Material and Methods: Fifty-four formalin fixed paraffin embedded (FFPE) OPSCC samples were analyzed for HPV DNA by a PCR-based bead-based multiplex-assay. Thirty-four samples were tested for HPV RNA and for overexpression of p16INK4 by immunohistochemistry. Twenty samples were evaluated by Competi- tive Allele-Specific Taqman PCR (CAST-PCR) for fibroblast growth factor receptor 3 protein (FGFR3) status. Results: A total of 33.3% (18/54) OPSCC samples were positive for HPV DNA. HPV16 was the most frequent type (30%, 16/54); followed by HPV18 (3.7%, 2/54); and 1 sample (1.8%) was positive for both HPV16 and 18. HPV18 E6*I was detected in a HPV18 DNA-positive oropharynx tumor. Four samples positive for HPV16 were also positive for p16INK4a. All the tested samples were negative for FGFR3.
Conclusions: The increased HPV16 prevalence is in line with similar studies and is a new confirmation that HPV16 is the most prevalent type in our country; supporting the potential benefit of prophylactic vaccines.Overall, there is no concordance between DNA and any of the two other analytes that are considered being markers of HPV-driven cancers. There is a need to explore novel screening strategies that could be broadly used in the clinical routine to initiate preventive measures.Keywords: HPV; cancer; biomarkers; early detection; survival
1.Introduction
Human papilloma virus (HPV) is recognized by the International Agency for Re- search on Cancer(IARC) as a biological agent involved in carcinogenesis as main co-factor. From all high-risk HPV types known to be involved in cervical cancer, only HPV16 was recognized as having an etiological role in oropharyngeal cancers and tonsil cancers [1]. HPV-driven head and neck squamous cell carcinomas (HPV-driven HNSCC) represent different malignancies, in comparison to cervical cancer. For cervical cancer types, for which HPV is recognized as a necessary cause, well-known screening methods (e.g., Pap test, and clinically validated high risk HPV DNA tests) are available, and together with the availability of the three vaccines (bi, tetra, and nonavalent), there is hope for cervical cancer elimination [2,3].
For HPV-driven HNSCC, clinically validated screening methods and therapeutic strategies are not yet established, as these categories of cancers are very heterogenous (oral cavity, oropharynx, pharynx,and larynx/hypopharynx).Information is still needed, regarding the understanding of the temporal and geographic differences in HPV-driven oropharyngeal squamous cellular cancers (OPSCC) incidence, as well as regarding the risk factors and if they change over time. A recent study from the USA revealed an increased proportion ofmiddle-aged patients with oropharyngeal cancer, which reflects the increase in HPV-related cancers [4]. The same increasing trend was published by the Dalianis HNSCC group, which detected a 70% HPV prevalence in TSCC/BOTSCC in Stockholm [5]. The epidemiology and natural history of oral HPV infection is important to be known. Similarly with the research realized for precancerous lesions in case of cervical cancer, it is necessaryto analyze the precursor lesions of HPV-driven OPSCC.For example, Haeggblom L et identified significant differences in protein expression for SPARC, psoriasin, type I collagen, and galectin-1 in both HPV+ and HPV− TSCC/BOTSCC, being one of the first studies which examined the differences in gene expression between dysplastic and invasive HPV+ and HPV−TSCC/BOTSCC [6].
Another biomarker is fibroblast growth factor receptor 3 (FGFR3), which has been identified as a potential target for therapy of HNSCCs. By NGS (next generation sequenc- ing), FGFR3 was found mutated in 15% of Norwegian HNSCC samples analyzed. This finding confirms the possibility of advanced personalized treatment, using targeted therapy against FFR3 [7]. Besides FGFR3, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) was found to be mostcommonly mutated in HNSCC. Both these gene mutations are used to identify new therapeutic targets andfurther our understanding of existing ones [8]. Erdafitinib and pemigatinib, two FGFR selective inhibitors targeting FGFR fusions, have been approved by the U.S. Food and Drug Administration (FDA) to treat patientswith urothelial cancer and cholangiocarcinoma, respectively [9,10], and there is hope that these inhibitors couldbe used for HNSCC also.
To identify patients at risk of severe evolution, testing for specific biomarkers is necessary, to relate to clinical practice. Recent studies from the USA note the changing ther- apeutic trends for advanced HPV-positive OPSCC: the percentage of HPV positive patients treated with surgery-based therapy decreasing by half,in the group of patients targeted by the study, during the period 2010–2016 [11]. Howard J et al., fromCambridge, evaluated, in a Cochrane review, the randomized controlled trials regarding the effects of de-intensified adjuvant (chemo)radiotherapy in comparison to standard adjuvant (chemo)radiotherapy in patients with resectable HPV-positive OPSCC, as it is known that HPV-positive OP- SCC patients are often younger and have a significantly improved survival relative to HPV-negative patients. The study will continue till 2023, to better analyze if acute and late toxicities associated with chemoradiotherapy are a significant burden for OPSCC patients, and if there are any adequate strategies that could decrease treatment-associated morbidity [12]. De-intensifying treatment trials in HPV-associated OPSCC was analyzed by researchers from Germanyand the USA [13,14].
The research in the OPSCC field has developed even more than this. Recently, a multicenter researchteam published their results in Nature, regarding the possibility of future therapeutic vaccines that target E6, which can attenuate viral response after HPV infections becomeestablished and might protect against HPV (+) OPSCC [15].
The estimated age-standardized incidence rates (ASR) in 2020, cervix uteri cancer, all ages, for Romania is 22.6, on the second place in European countries, after Montenegro, and the estimated ASR in 2020, for oropharynx, lip, oral cavity cancers, males, all ages, Europe is 18.0, being first place in European countries[16], which underlines the fact that HNSCC represent an important health issue in our country. We have experience in testing HNSCC patients from our region, from fresh tumor and from FFPE samples, but there is aneed for more comprehensive studies of HNSCC in our area, to analyze the risk factors and the clinical utility[17–20].
AIM: To analyze the HPV driven cases in selected OPSCC patients, using specific biomarkers, inrelation to an updated scientific literature review.
2.Materials and Methods
The patients assessed in this study belong to two previous study cohorts, the HPV- AHEAD study realized in collaboration with IARC-Lyon, and a collaborative study between the University of Medicine and Pharmacy “Grigore T. Popa”, Iasi, Romania and Karolinska Institute-Stockholm. Fifty-four HNSCC patients were selected from the Departments of Oral and Maxillofacial Surgery, Otorhinolaryngology, “Grigore T. Popa” University of Medicine and Pharmacy (Ias, i, Romania), from January 2010 to October 2017. The formalin- fixed, paraffin-embedded (FFPE) HNSCC blocks included squamous cell carcinoma of the oropharynx (International Classification of Diseases for Oncology (ICD-O) C01—base of tongue, C02.4—lingual tonsil, C09—tonsil, C10—oropharynx) and were retrieved from the hospital archives. The OPSCC samples were collected from patients with a median age of 54 years (limits: 41–79), from which 9% (5/54) were women, 54% (29/54) declared being smokers, and 21% (11/54) declared being chronic alcohol users. The education ofthe patients was high school or professional school. All patients were diagnosed with keratinizing or non-keratinizing squamous cell carcinomas. One patient with base of tongue of tongue cancer was treatedpreviously (2014) with radiotherapy for larynx carcinoma.
Ethical clearance for the investigations reported in this study was obtained from the Institutional Ethical Committee of the “Grigore T. Popa” University of Medicine and Pharmacy, Ias, i, Romania, reference number 7150/2014 and 3953/2018 respectively. The study implied the use of archival material only, and it did not envisage any contact with the patients or to disclosure any personal data files. Adequate measures to ensure data protection, confidentiality, patients’ privacy, and anonymization were considered. No informedconsent was available. All data were fully anonymized before access.
Preparation of paraffin sections and DNA extraction: The FFPE blocks were each sec- tioned into 31 sections, following the HPV-AHEAD protocol; three were used for histology analysis (sections S1, S10, and S31), two for p16 immunohistochemistry (S2 and S9), and S11 ± S30 were archived for future independentstudies. For DNA and RNA and RNA analysis, sections S3 ± S5 and S6 ± S8 were collected in two separate vials. In order to ensure the risk of cross-contamination was minimal, each FFPE block was sectioned using a different blade. Ethanol 70% and DNA Away (Dutscher, Brumath, France) were used to clean the microtome after each block. After every 10th tissue FFPE block, an empty paraffin block was sectioned and processed to control for DNA cross-contamination. For DNA extraction, the paraffin tissue sections were incubated overnight in a digestion buffer (10 mM Tris/HCl pH 7.4, 0.5 mg/mL proteinase K, and 0.4% Tween 20). Nucleic acids extraction was performed from FFPE OPSCC sections according to QIAamp DNA FFPETissue Procedure: remove paraffin from sample, lyse with proteinase K, heat, bind DNA, wash, and elute theready to use DNA in 50 µL buffer.
HPV DNA genotyping: Type-specific multiplex genotyping (TS-MPG) assay, a method combining multiplex PCR and bead-based Luminex technology (Luminex Corporation, Austin, TX, USA), was used to determine HPV DNA positivity [21]. This assay is able to detect 19 HR (HPV16, 18, 31, 33, 35,39, 45, 51, 52, 56, 58, and 59) or probable high-risk (HPV26, 53, 66, 68a and b, 70, 73, 82) and two low-risk HPV types (HPV6 and 11). Cellular beta-globin gene was used to control for DNA quality. Following PCR amplification, a 10 µL sample of each reaction mixture was used for multiplex HPV genotyping by Luminex technology (Luminex Corporation, Austin, TX, USA)[22].
HPV RNA analysis: Total RNA was purified using the Pure Link FFPE Total RNA Isolation Kit (Invitrogen, Carlsbad, CA, USA) from three pooled sections of the same tissue block. RT-PCR analysis was carried out using the QuantiTect Virus Kit (Qiagen, Hilden, Germany), in a total volume of 25 µL containing5 µL of 5xQuantiTect Virus Mastermix, 0.25 µL of 100xQuantiTect Virus RT Mix, 0.4 µM of each oligonucleotide, and 1 µL RNA.
The HPV type-specific E6*I mRNA assay was used to detect the viral transcripts. This analysis amplifies a 65 ± 75 base pair sequence of HPV and an 81 base pair sequence of ubiquitin C cDNA. Biotinylated amplification products are hybridized to ubC and HPV type-specific probes representing splice junction sequences on Luminex beads, followed by staining with streptavidin-phycoerythrin, and quantified in a Luminex analyzer. Byusing a splice product sequence as the detection probe, absolute specificity for RNA is ensured, eliminating the risk of false positivity of the assays using unspliced RNA, which comes from residual viral DNA and RNAfrom the preparation [23].
Expression of p16 was assessed by IHC in FFPE sections using the CINtec p16 Histol- ogy kit (Roche mtm laboratories AG, Mannheim, Germany) according to the instructions of the manufacturer. Positive p16expression was defined as diffuse nuclear and cytoplasmic staining in 70% or more of the tumor cells. Thevalidity of the p16 IHC staining result was assessed by evaluating the presence of p16 internal control staining. Discrepant results were re-checked by a pathologist, and the final classification of the staining was based on themajority consensus of the working group [24].
Competitive allele-specific Taqman PCR (CAST-PCR): FGFR3 mutations were de- tected by Competitive Allele-Specific Taqman® PCR technology (Thermo Fischer Scientific, Waltham, MA, USA), an assay performed in 384-well plates, in a total volume of 10 µL com- prising 5 µL 2X Taqman GenotypingMastermix, 0.2 µL 50X Exogenous IPC template DNA, 1 µL 10X Exogenous IPC mix, 1 µL Mutation Detection Assay, 1.8 µL deionized water and 20 ng DNA (in 1 µL). Applied Biosystems 7900HT Fast Real-Time PCR System was used with the following set of conditions: 95 ◦C, 10 min followed by 5 cycles at 92 ◦C, 15 s and 58 ◦C, 1 min and 40 cycles at 92 ◦Cfor 15 s, and 60 ◦C for 1 min. SDS 2.3 Software was used to analyze the PCR result, together with MutationDetection Software 2.0 (Thermo Fischer Scientific, Waltham, MA, USA). Ct was determined for exogenous IPC (Internal Passive Control) reagentsadded to each reaction to evaluate PCR failure or inhibition in a reaction. The Mutation Detection Assays Hs00000811_mu, Hs00000812_mu, Hs00001342_mu were used to detect variants p.R248C, p.S249C and p.K650Q in FGFR3 gene respectively, and Hs00001015_rf reference assay was used to identify the presence of wild-type FGFR3. All these assays were previously described in detail [19,20].
3.Results
HPV DNA genotyping: 18/54 (33.4%) tissue samples were DNA/HPV positive, out of which 16/54(29.6%) were HPV16 positive, 2/54 were HPV18 positive, and one (1.85%) was detected as having doubleHPV16 and HPV18 infection.
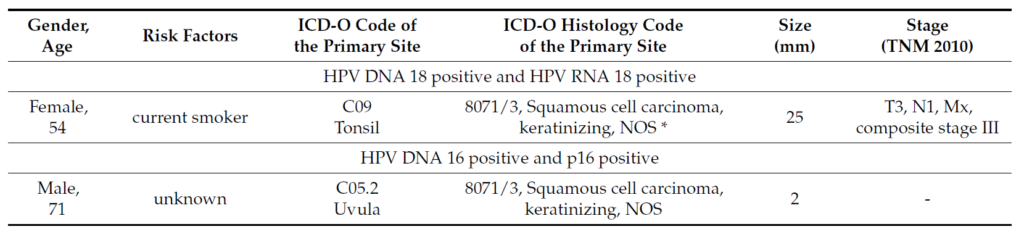
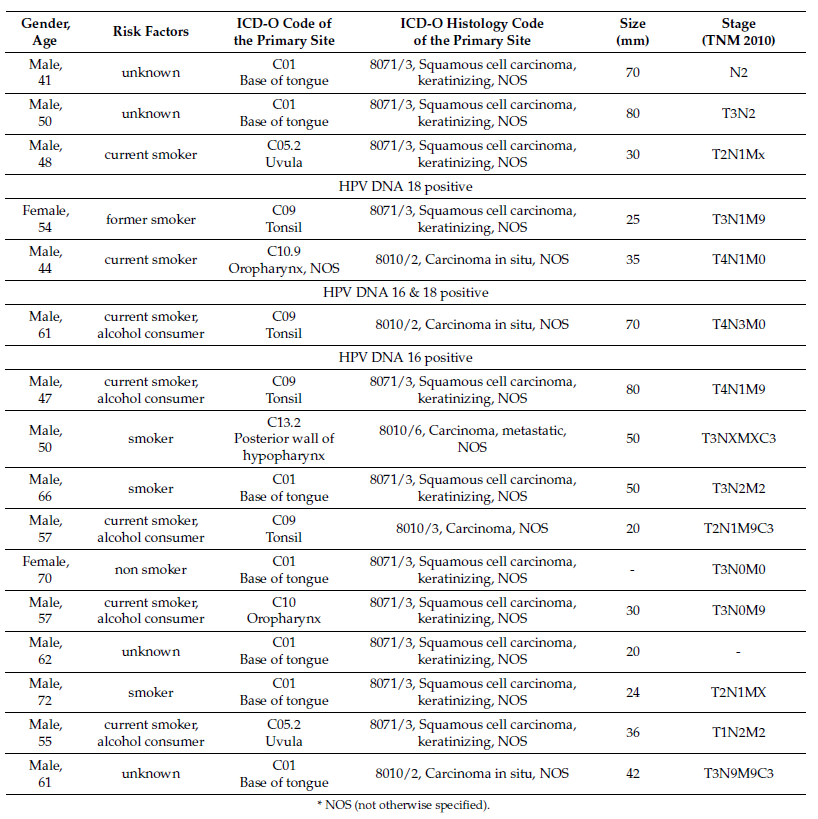
HPV RNA analysis: 1/34 (1.96%) was positive for HPV18 E6*I and for HPV DNA 18. Expression of p16was assessed by IHC: 4/34 (11.76%) HPV DNA 16 were also positive for p16INK4a (Figure 1). Acontinuous, diffuse staining for p16 within the cancer area of the tissue sections was considered positive, whilea focal staining or no staining were considered negative. The cut-off of p16 positivity was considered adiffuse staining corresponding to 70% of p16 positive tumor cells [13]. IHC slides were evaluated by onepathologist blinded to any other clinical information or HPV DNA or RNA status. The validity of the p16IHC staining result was assessed by evaluating the presence of p16 internal control staining. The clinicalcharacteristics for the p16 positive RNA and DNA HPV positive cases can be seen in Table 1.
Competitive allele-specific Taqman PCR: all the samples tested by CAST PCR for FGFR3 mutation were negative.
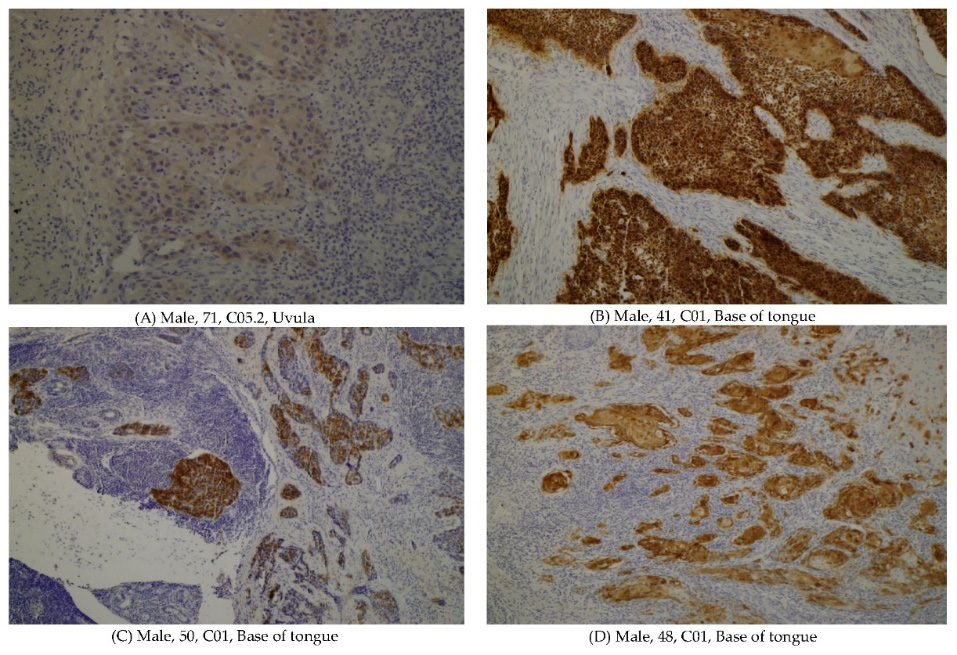
Figure 1. The p16 positive oropharyngeal cancers. (A) Histology sample from a 71 y.o. male patient, unknown risk factors, C05.2 Uvula, 8071/3 Squamous cell carcinoma, keratinizing, NOS. (B) Histology sample from a 41 y.o. male patient, unknown risk factors, C01 Base of tongue, 8071/3 Squamous cell carcinoma, keratinizing, NOS. (C) Histology sample from a 50 y.o. male patient, unknown risk factors, C01 Base of tongue, 8071/3 Squamous cell carcinoma, keratinizing, NOS. (D) Histology sample from a 48 y.o. male patient, current smoker, C05.2 Uvula, 8071/3 Squamous cell carcinoma, keratinizing, NOS.
Table 1. Descriptive characteristics of all HPV OPSCC positive cases.
4.Discussion
It is already well known that mucosal HR HPV-types (HPV16) are involved in OP- SCC [25], and thecontribution of HR HPV to the carcinogenesis of HNSCC is different by geographical region [26]. HPV-positive OPSCC was defined as positivity to HPV DNA and p16 IHC, and HPV-driven OPSCC was defined as being HPV DNA and RNA posi- tive [27–29]. The HPV-positive OPSCC refers to carcinomas of the oropharynx presumed to be associated with HPV, mostly based on positivity to HPV DNA and p16 IHC [27].
In our country, there is a known high burden of cervical cancer, and it is expected to detect the same highprevalence of HPV in HNSCC, as it is in cervical cancer. Our detected HPV-positive OPSCC prevalence was4/54 (7.4%) HPV-positive patients and 1/54 (1.8%) patient with an HPV-driven HNSCC case. Our results OPSCC HNSCC originate from samples such as tonsil,base of tongue, and uvula, in line with other studies [20].
Potential reasons for the generally low rates of positive agreement between p16 and RNA with HPV DNApositivity might be several technical issues, such as fixation artifacts and over-fixation that may impact both p16 IHC and RNA analysis out of FFPE. Another reason can be the potentially low rate of HPV-driven carcinogenesis in this Romanian cohort, where most HPV DNA positive cases may not be, in reality, cases where HPV drives the carcinogenesis (i.e., detection of clinically irrelevant HPV DNA infections in the tissue). The high percentage of smokers (54%) and even chronic alcohol users (21%), known as risk factors,could be another explanation [17,19].
Moreover, it is interesting that, in our samples, we did not detect any FGFR mutation, in comparison to the Swedish cohort in which 11/109 HPV+ TSCC/BOTSCC patients exhibited FGFR3 mutations. For the Swedish patients, the high FGFR3 expression among patients with wild-type FGFR3 was correlated to better disease-free survival [20].
- 4.1. OPSCC HPV DNA Positive, p16, Risk Factors
The pattern of OPSCC cancers is a research topic for many authors. We carried out an analysis of therecently published articles which are referring to HPV-positive OPSCC.
A research team from the Johns Hopkins University analyzed a comprehensive num- ber (more than 13,000) of these types of cancers, HPV-positive, for twelve years. The authors found that the BOTSCC and TSCC are different from an epidemiological point of view, and they identified risk factors such as: age over 70, white race, male gender, and tonsillectomy [30]. Authors from Belgium analyzed 131 patients over a period 10 years (2007–2017) and concluded that p16 expression was associated with a better evolution of the disease. Besides p16, the authors included smoking and chronic alcohol use in their prognostic significance score [31].Smoking was also taken in consideration as a risk factor for survival of HPV-positive oropharyngeal cancers, by ateam from the USA. The authors found that for a smoking status of more than 10 pack/year, the effect on five-year overall survival was significant, and therefore they considered that smoking should be taken inconsideration by The American Joint Committee on Cancer (AJCC), in the staging system of HPV-positive oropharyngeal cancer [32]. Gale N et al., used a recently developed HR-HPV (high-risk human papillomaviruses) mRNA silver in situ hybridization (SISH) assay for detecting HPV16, HPV18, and HPV33in HPV DNA positive oropharyngeal cancers, as it is known that the transcriptional activity of HR-HPV within oropharyngeal squamous cell carcinomas has been linked to improved survival of the patients. The authors found an 80% sensitivity of this new method, which supports the need for further studies using this assay [33]. Another survival marker of HPV-positive OPSCC was considered to be HPV16 viral load by Japanese authors, as it is also thought to be for cervical cancer [34]. In Brazilian HPV+ OPSCC patients, Santos Carvalho R et al., identified tumor staging, tumor localization, smoking, level of education, and gender as prognosis factors. The authors suggest that these factors could be used for a better stratification andmanagement of this category of patients [35]. A research team from Lyon used p16/p53 algorithm to predictHPV tumor status in OPSCC, and authors from Germany concluded that tobacco, alcohol abuse, age, and gender should be added to the current UICC (The Union for International Cancer Control) staging system to improve the risk stratification in HPV+ OPSCC [13,36]. Korsten LHA et al. from Amsterdam evaluated the health-related quality of life for HPV-positive OPSCC patients. For the very pragmatic question, “Does HPVstatus matter?”, the authors found that HPV-positive OPSCC patients had a better score before treatment,recovered faster, and had a different course of emotional functioning, in comparison to HPV-negative patients[37]. A research team from Denmark investigated, in a review study, the impact of specific HPV genotypes on survival in HPV+ OPSCC and identified a favorable prognosis among patients with HPV16 OPSCC compared with HR non-HPV16 OPSCC [38].
- 4.2. OPSCC HPV DNA Positive and HPV RNA Positive
Current HPV detection methods in OPSCC have varying sensitivity and specificity. Many authors included HPV RNA testing in their studies. Lu XJD et al. analyzed, in Canada, more than 200 patientsdiagnosed with OPSCC, using RNA in situ hybridization (ISH) for HPV16 and 18, beside other biomarkers. For the samples that they tested, the authors detected a higher sensitivity for HPV testing in comparison to p16, and they concluded that HPV RNA ISH testing could be used as a routine analysis [39]. Another team of researchers from the USA used mRNA in situ hybridization (mRNA ISH) for HPV detection in OPSCC anddetected a sensitivity and specificity of 100% for both in comparison to p16 analysis, which are ideal parameters for any laboratory test [40]. Zito Marino F et al. simultaneously tested HPV RNA ISH and p16 IHC on the same slide, for both cervical cancer and OPSCC samples. The results obtained by this doubletesting was similar with the results previously obtained by classical p16 and HPV RNA testing. The authors areunderlying the ease of applicability of this assay on FFPE samples, with complete automation, and possibility forimplementation in clinical laboratories [41]. Researchers from Finland, with recognized experience in the HNSCC field, analyzed OPSCC samples by different methods, including HPV mRNA ISH. From all assays used, the ISH for high-risk HPV E6/E7 mRNA had the highest sensitivity (93%). The authors correlated thecharacteristics of this test with the necessity of de-escalating the therapy for HPV-positive tumors, at safe doses to maintain the favorable outcome [42]. Shinn JR et al. developed a study starting from the lack ofconcordance between p16 and HPV RNA testing. Despite this, the authors carried out a large contemporary retrospective study (nearly 500 patients), in the USA, in which they analyzed samples of OPSCC by these twoassays. They concluded that it would be more efficient to confirm cases by HPV mRNA testing, as theyidentified that p16 status is not consistent with tumor morphology [43]. A very interesting study evaluated the prognostic utility, in HPV16-associated OPSCC, of HPV16 E6/E7 expression in circulating tumor cells. This expression was correlated with HPV16 DNA status and with p16. The authors concluded that this non-invasiveliquid biopsy test for circulating tumor cells together with UICC staging could improve risk discrimination at the onset of disease [44]. This study is based on the same principle of microchimerism analyzed in other tumors and clinical circumstances [45]. A team of researchers from Ireland found that using the HPV RNA-ISHmethod was a more specific test in confirming HPV status, in comparison to the p16 assay [46].
- 4.3. HPV DNA + HPV RNA + p16INK4a Over-Expression
A comprehensive study of HNSCC, the HPV-AHEAD project, was realized by an international team of researchers, under the guidance of Infections and Cancer Biology Group, International Agency for Research onCancer, Lyon, France. The algorithm used in this study was a very strict and rigorous one: FFPE HNSCCsamples were analyzed using HPV DNA testing, HPV RNA testing, and p16 analysis. Archived HNSCC tissuesamples from 772 patients from Belgium, 696 from Italy, 364 from the central region of India, and 189 from North-East Romania were tested using this algorithm. In all the four HPV-AHEAD studies, the highest rate of HPV DNA prevalence was detected for OPSCCs, with similar values (36.4% in Belgium, 40.4% in Italy, 18.9% in India, and 50% in Romania). HPV16 was the most prevalent type in all the samples analyzed by these studies. HPV-driven HNSCCs were defined by the presence of both viral DNA and RNA, and the highest prevalence of this double positivity was also found in OPSCC samples [19,47–49].
- 4.4. HNSCC HPV Positive PIK3CA and FGFR Positive
Seiwert TY et al., evaluated the genetic differences between HPV-positive and HPV- negative HNSCC, by sequencing carried out at the Institute for Genomics and Systems Biology, University of Chicago. The authors found that HPV-positive tumors showed unique mutations in DDX3X, FGFR2/3, and aberrations in PIK3CA and KRAS. The mu- tation of the FGFR and PI3K genes were considered by the authors targetable genes for therapy [50]. As fibroblast growth factor receptor family member proteins (FGFR1-4) have been identified as promising novel therapeutic targets and prognostic markers in a wide spectrum of solid tumors, researchersfrom the Netherlands Cancer Institute analyzed the protein expression of FGFR1-4 by immunohistochemistry, on tissue microarrays. FGFR proteins were highly expressed in oral cavity (39–64%) and oropharyngeal squamous cell carcinoma (63–79%); therefore, the authors considered that FGFRs may be potential targetsfor therapy, in order to optimize the survival of this category of HNSCC patients [51]. Even though thetherapeutic benefit of FGFR inhibitors is already known, the appearance of resistance is also a known possibility. The same research team from the Netherlands evaluated, one year later, the possibility to avoid or delay resistance, by using AZD4547 and gefitinib in FGFR inhibitor resistant HNSCC patients [52]. Bersani et al. from Karolinska Institute analyzed by NGS, HPV+ TSCC/BOTSSCC and HPV+ HNSCCUP and HPV−TSCC/BOTSCC samples. The authors found that the mutation of PIK3CA was most fre- quently observed in HPV-positive tumors, and FGFR3 mutation was associated with severe prognosis [53]. Researchers from Korea also studied HNSCC and their correspondent blood samples by NGS, and they found that the most frequent mutations were for TP53, CDKN2A, CCND1, and PIK3CA. This team concluded that the implementation of precision medicine in HNSCC is feasible and requires clinical trials to assess the efficiency of molecu- lar targeted agents [54]. Bersani C et al. developed a special assay to detect FGFR mutation,Competitive Allele-Specific Taqman PCR (CAST-PCR). This method was cost efficient, in comparison to the previous NGS and useful to correlate the expression of FGFR3 with disease free survival [20]. Sablin MP et al. tested by Real Time PCR 42 genes coding for major druggable proteins, from selected HNSCC patients. The authors identified that the most significantly overexpressed genes involved cell cycle regulation tyrosine kinasereceptors, angiogenesis, and immune system and PIK3CA expression, and they considered that these genes aredruggable [55].
Similar research studies have also been made by other authors regarding molecular profiling of HNSCC: Feldman R et al. identified TP53 and PIK3CA mutations among HPV-positive or HPV-negative patients [56]; and Gillison ML et al. detected ZNF750, PIK3CA, and EP300 mutations as candidate driver events in HPV-positive cancers [57]. Understanding the mutational landscape of HNSCC has opened the path of evaluating optimized therapeutic approaches to manage this group of diseases. Additional insight into the molecular subtypes of HNSCC and its specific subsites will further drive improved strategies to stratify and treat patients with this debilitating disease [58]. Given these detected gene mutations in HNSCC, some authors evaluated targeted therapy in corre- sponding cell lines, such as, for example, evaluating the sensitivity of TSCC/BOTSCC cell lines to PIK3CA. Food and Drug Administration (FDA) approved the drugs alpelisib(BYL719) and erdafitinib (JNJ-42756493) alone and in combination with cisplatin or doc- etaxel [59,60].BYL719 was tested by other authors in HNSCC cell lines therapy [61]. As a logical continuation after these preclinical studies, a two-phase II clinical trials were carried out regarding deintesification of CRT for PIK3CA mutations in HPV-associated OPSCC patients. Patients with wild-type-PIK3CA had statistically significantly higher 3-year disease-free survival than PIK3CA-mutant patients [62]. In another clinical trial which analyzed HNSCC patients, Geiger JL et al. concluded that everolimus, an mTOR inhibitor, was not active asmonotherapy in unselected patients with recurrent/metastatic HNSCC [63].
The scientific literature cited above presents the results of studies realized using one or more assays toevaluate the role of HPV in OPSCC. This brief review proves the difficulty elements in the research of this issue, focusing on the possible detection, follow up and targeted therapy of HPV positive OPSCC, and the limitations of current approaches [64]. The above review of very recent research supports the continuation of this study in our area, as it is known that a virus-induced tumor could be efficiently prevented by a pan-genderHPV vaccination [65] or treated by oncolytic viral therapy and/or by targeted therapy, such as the newlyemerged immune checkpoint inhibitors (PD-1/PD-L1 pathway) [66].
Our study has its limitations. The small number of analyzed patients and the detected results raise more questions and concerns regarding the HPV status in our OPSCC cancers, the most adequate screening method,and if the targeted FGFR and PI3K therapy could be useful for these categories of patients. At the moment, thisis one of the few studies of this kind in our area. For the use of this research to materialize, this study should becontinued with a very well-organized cohort study, to be able to select and analyze, using specific biomarkers,the OPSCC patients with the highest optimal response to targeted therapy.
5.Conclusions
The increased HPV prevalence (HPV16) shows one more time the potential benefit of prophylactic vaccines. We have detected a higher concordance between HPV DNA and p16 than between HPV DNA andRNA, even though not statistically significant. Overall, there is no concordance between DNA and any of thetwo other analytes that are considered as being markers of HPV-driven cancers. There is a need for prospective studies to describe risk factors and the natural history of oral HPV infections and to explore novel screeningstrategies that could be broadly used in the clinical routine to initiate preventive and therapeutic measures.
Author Contributions: Conceptualization: R.G.U.; methodology, R.G.U. and L.S.I.; resources, S.E.G., I.A.S., B.M., M.D., I.L., E.P.-A., N.G., V.V.C., D.O.P., P.T. and M.A.V.; writing—original draft prepara- tion, R.G.U.; writing—review and editing, R.G.U., C.D. and L.S.I. All authors have read and agreed to the published version of the manuscript.
Funding: The study was undertaken by R.G. Ursu while hosted as Visiting Scientist by the Interna- tional Agency for Research on Cancer, Lyon, France, and at Karolinska Institute, Stockholm. Scientific research funded by “Grigore T. Popa” University of Medicine and Pharmacy, Ias, i, research contract 6983 from 21.04.2020. This study was supportedby the European Commission, grant HPV-AHEAD (FP7-HEALTH-2011-282562) to MT.
Institutional Review Board Statement: The study was conducted according to the guidelines of the Declaration of Helsinki and approved by Ethics Committee of “Grigore T. Popa” University of Medicine and Pharmacy, Iasi(7150/2015 and 3953/2018, respectively).
Informed Consent Statement: Informed consent was not necessary for this retrospective study.
Data Availability Statement: Data are contained within the article.
Acknowledgments: We are grateful to Tina Dalianis and to Massimo Tommasino who gave permis- sion to RamonaUrsu to test these tumor samples at Karolinska Institute, during successive research mobilities, and at IARC, during herinternship program as Visiting Scientist at Infections and Cancer Biology Group, International Agency for Research on Cancer, Lyon, France. Part of these data have been presented at the 4th Workshop on Emerging Issues in Oncogenic Virus Research 2016, Manduria, Italia and at EUROGIN 2019, Monaco.
Conflicts of Interest: The authors declare no conflict of interest.
References
- International Agency for Research on Cancer (Ed.) A Review of Human Carcinogens: Biological Agents; IARC Monographs of the Evaluation ofCarcinogenic Risks to Humans; IARC: Lyon, France, 2012; ISBN 978-92-832-1319-2.
- Dillner, J.; Elfström, K.M.; Baussano, I. Prospects for Accelerated Elimination of Cervical Cancer. Prev. Med. 2021, 153, 106827.[CrossRef] [PubMed]
- Hall, M.T.; Simms, K.T.; Lew, J.-B.; Smith, M.A.; Brotherton, J.M.; Saville, M.; Frazer, I.H.; Canfell, K. The Projected Timeframe until CervicalCancer Elimination in Australia: A Modelling Study. Lancet Public Health 2019, 4, e19–e27. [CrossRef]
- Bajpai, S.; Zhang, N.; Lott, D.G. Tracking Changes in Age Distribution of Head and Neck Cancer in the United States from 1975 to 2016. Clin.Otolaryngol. 2021, 46, 1205–1212. [CrossRef] [PubMed]
- Haeggblom, L.; Attoff, T.; Yu, J.; Holzhauser, S.; Vlastos, A.; Mirzae, L.; Ährlund-Richter, A.; Munck-Wikland, E.; Marklund, L.; Hammarstedt-Nordenvall, L.; et al. Changes in Incidence and Prevalence of Human Papillomavirus in Tonsillar and Base of Tongue Cancer during 2000–2016 in theStockholm Region and Sweden. Head Neck 2019, 41, 1583–1590. [CrossRef] [PubMed]
- Haeggblom, L.; Ährlund-Richter, A.; Mirzaie, L.; Farrajota Neves da Silva, P.; Ursu, R.G.; Ramqvist, T.; Näsman, A. Differences in Gene Expression between High-grade Dysplasia and Invasive HPV+ and HPV− Tonsillar and Base of Tongue Cancer. Cancer Med. 2019, 8, 6221–6232.[CrossRef]
- Dongre, H.N.; Haave, H.; Fromreide, S.; Erland, F.A.; Moe, S.E.E.; Dhayalan, S.M.; Riis, R.K.; Sapkota, D.; Costea, D.E.; Aarstad, H.J.; et al.Targeted Next-Generation Sequencing of Cancer-Related Genes in a Norwegian Patient Cohort With Head and Neck Squamous Cell Carcinoma Reveals Novel Actionable Mutations and Correlations With Pathological Parameters. Front. Oncol. 2021, 11, 734134. [CrossRef] [PubMed]
- Swaney, D.L.; Ramms, D.J.; Wang, Z.; Park, J.; Goto, Y.; Soucheray, M.; Bhola, N.; Kim, K.; Zheng, F.; Zeng, Y.; et al. A Protein Network Map of Head and Neck Cancer Reveals PIK3CA Mutant Drug Sensitivity. Science 2021, 374, eabf2911. [CrossRef] [PubMed]
- Zengin, Z.B.; Chehrazi-Raffle, A.; Salgia, N.J.; Muddasani, R.; Ali, S.; Meza, L.; Pal, S.K. Targeted Therapies: Expanding the Role of FGFR3Inhibition in Urothelial Carcinoma. Urol. Oncol. Semin. Orig. Investig. 2021, in press. [CrossRef]
- Chen, L.; Zhang, Y.; Yin, L.; Cai, B.; Huang, P.; Li, X.; Liang, G. Fibroblast Growth Factor Receptor Fusions in Cancer: Opportunities andChallenges. J. Exp. Clin. Cancer Res. 2021, 40, 345. [CrossRef]
- Mirza, F.A.; Johnson, C.Z.; Byrd, J.K.; Albergotti, W.G. Treatment Trends for Advanced Oropharyngeal Squamous Cell Carcinoma in the Era ofHuman Papillomavirus. Head Neck 2021, 43, 3476–3492. [CrossRef] [PubMed]
- Howard, J.; Dwivedi, R.C.; Masterson, L.; Kothari, P.; Quon, H.; Holsinger, F.C. De-Intensified Adjuvant (Chemo)Radiotherapy versus StandardAdjuvant Chemoradiotherapy Post Transoral Minimally Invasive Surgery for Resectable HPV-Positive Oropha- ryngeal Carcinoma. Cochrane DatabaseSyst. Rev. 2018, 12, CD012939. [CrossRef] [PubMed]
- Oberste, M.; Riders, A.; Abbaspour, B.; Kerschke, L.; Beule, A.G.; Rudack, C. Improvement of Patient Stratification in Human Papilloma Virus-associated Oropharyngeal Squamous Cell Carcinoma by Defining a Multivariable Risk Score. Head Neck 2021, 43, 3314–3323. [CrossRef]
- Lu, D.J.; Luu, M.; Nguyen, A.T.; Scher, K.S.; Clair, J.M.-S.; Mita, A.; Shiao, S.L.; Ho, A.S.; Zumsteg, Z.S. Survival Outcomes with Concomitant Chemoradiotherapy in Older Adults with Oropharyngeal Carcinoma in an Era of Increasing Human Papillomavirus (HPV) Prevalence. Oral Oncol.2019, 99, 104472. [CrossRef] [PubMed]
- Ferreiro-Iglesias,A.; McKay, J.D.; Brenner, N.; Virani, S.; Lesseur, C.; Gaborieau, V.; Ness, A.R.; Hung, R.J.; Liu, G.; Diergaarde, B.; et al. Germline Determinants of Humoral Immune Response to HPV-16 Protect against Oropharyngeal Cancer. Nat. Commun. 2021, 12, 5945. [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: CancerToday. Available online: http://gco.iarc.fr/today/home (accessed on 2 November 2021).
- Ursu, R.G.; Luchian, I.; Ghetu, N.; Costan, V.V.; Stamatin, O.; Palade, O.D.; Damian, C.; Iancu, L.S.; Porumb-Andrese, E. Emerging OncogenicViruses in Head and Neck Cancers from Romanian Patients. Appl. Sci. 2021, 11, 9356. [CrossRef]
- Ursu, R.G.; Ivanov, I.; Popescu, E.; Costan, V.; Stamatin, O.; Ghetu, N.; Palade, D.; Mirtu, C.; Andrese, E.; Danciu, M.; et al. Human papillomavirus genotyping in fresh head and neck tumors—Our first experience. Med.-Surg. J. 2015, 119, 676–680.
- Ursu, R.G.; Danciu, M.; Spiridon, I.A.; Ridder, R.; Rehm, S.; Maffini, F.; McKay-Chopin, S.; Carreira, C.; Lucas, E.; Costan, V.-V.; et al. Role of Mucosal High-Risk Human Papillomavirus Types in Head and Neck Cancers in Romania. PLoS ONE 2018, 13, e0199663. [CrossRef]
- Bersani, C.; Haeggblom, L.; Ursu, R.G.; Giusca, S.E.; Marklund, L.; Ramqvist, T.; Näsman, A.; Dalianis, T. Overexpression of FGFR3 in HPV-Positive Tonsillar and Base of Tongue Cancer Is Correlated to Outcome. Anticancer Res. 2018, 38, 4683–4690. [CrossRef] [PubMed]
- Schmitt, M.; Dondog, B.; Waterboer, T.; Pawlita, M.; Tommasino, M.; Gheit, T. Abundance of Multiple High-Risk Human Papillomavirus (HPV)Infections Found in Cervical Cells Analyzed by Use of an Ultrasensitive HPV Genotyping Assay. J. Clin. Microbiol. 2010, 48, 143–149. [CrossRef]
- Gheit, T.; Abedi-Ardekani, B.; Carreira, C.; Missad, C.G.; Tommasino, M.; Torrente, M.C. Comprehensive Analysis of HPV Expression inLaryngeal Squamous Cell Carcinoma. J. Med. Virol. 2014, 86, 642–646. [CrossRef] [PubMed]
- Halec, G.; Schmitt, M.; Dondog, B.; Sharkhuu, E.; Wentzensen, N.; Gheit, T.; Tommasino, M.; Kommoss, F.; Bosch, F.X.; Franceschi, S.; et al. Biological Activity of Probable/Possible High-Risk Human Papillomavirus Types in Cervical Cancer. Int. J. Cancer 2013, 132, 63–71. [CrossRef][PubMed]
- Mena, M.; Lloveras, B.; Tous, S.; Bogers, J.; Maffini, F.; Gangane, N.; Kumar, R.V.; Somanathan, T.; Lucas, E.; Anantharaman, D.; et al. Developmentand Validation of a Protocol for Optimizing the Use of Paraffin Blocks in Molecular Epidemiological Studies: The Example from the HPV-AHEADStudy. PLoS ONE 2017, 12, e0184520. [CrossRef] [PubMed]
- Castellsagué, X.; Alemany, L.; Quer, M.; Halec, G.; Quirós, B.; Tous, S.; Clavero, O.; Alòs, L.; Biegner, T.; Szafarowski, T.; et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J. Natl. Cancer Inst. 2016, 108, djv403.[CrossRef]
- Ndiaye, C.; Mena, M.; Alemany, L.; Arbyn, M.; Castellsagué, X.; Laporte, L.; Bosch, F.X.; de Sanjosé, S.; Trottier, H. HPV DNA, E6/E7 MRNA, and P16INK4a Detection in Head and Neck Cancers: A Systematic Review and Meta-Analysis. Lancet Oncol. 2014, 15, 1319–1331. [CrossRef]
- Boscolo-Rizzo, P.; Pawlita, M.; Holzinger, D. From HPV-Positive towards HPV-Driven Oropharyngeal Squamous Cell Carcinomas.Cancer Treat Rev. 2016, 42, 24–29. [CrossRef]
- Rietbergen, M.M.; Leemans, C.R.; Bloemena, E.; Heideman, D.A.M.; Braakhuis, B.J.M.; Hesselink, A.T.; Witte, B.I.; Baatenburg de Jong, R.J.;Meijer, C.J.L.M.; Snijders, P.J.F.; et al. Increasing Prevalence Rates of HPV Attributable Oropharyngeal Squamous Cell Carcinomas in the Netherlandsas Assessed by a Validated Test Algorithm. Int. J. Cancer 2013, 132, 1565–1571. [CrossRef]
- Smeets, S.J.; Hesselink, A.T.; Speel, E.-J.M.; Haesevoets, A.; Snijders, P.J.F.; Pawlita, M.; Meijer, C.J.L.M.; Braakhuis, B.J.M.; Leemans, C.R.; Brakenhoff, R.H. A Novel Algorithm for Reliable Detection of Human Papillomavirus in Paraffin Embedded Head and Neck Cancer Specimen. Int. J.Cancer 2007, 121, 2465–2472. [CrossRef]
- Windon, M.J.; Fakhry, C.; Margalit, D.N.; Dey, T.; Rettig, E.M. Epidemiologic Distinctions between Base of Tongue and Tonsil OropharyngealCarcinomas. Head Neck 2021, 43, 3076–3085. [CrossRef]
- Bouland, C.; Dequanter, D.; Lechien, J.R.; Hanssens, C.; De Saint Aubain, N.; Digonnet, A.; Javadian, R.; Yanni, A.; Rodriguez, A.; Loeb, I.; et al. Prognostic Significance of a Scoring System Combining P16, Smoking, and Drinking Status in a Series of 131 Patients with Oropharyngeal Cancers.Int. J. Otolaryngol. 2021, 2021, 8020826. [CrossRef] [PubMed]
- Horwich, P.; Gundale, A.; Patin, S.; Flores, J.; Moore Medlin, T.; Chang, B.A.; Nathan, C.-A.O. Impact of Smoking on Stage-Specific Survival in Human Papilloma Virus-Associated Oropharyngeal Squamous Cell Carcinoma. Head Neck 2021, 43, 2698–2704. [CrossRef]
- Gale, N.; Poljak, M.; Volavšek, M.; Hošnjak, L.; Velkavrh, D.; Bolha, L.; Komloš, K.F.; Strojan, P.; Anicˇin, A.; Zidar, N. Usefulness of High-Risk Human Papillomavirus MRNA Silver in Situ Hybridization Diagnostic Assay in Oropharyngeal Squamous Cell Carcinomas. Pathol. Res. Pract.2021, 226, 153585. [CrossRef]
- Hashida, Y.; Higuchi, T.; Matsumoto, S.; Iguchi, M.; Murakami, I.; Hyodo, M.; Daibata, M. Prognostic Significance of Human Papillomavirus 16Viral Load Level in Patients with Oropharyngeal Cancer. Cancer Sci. 2021, 112, 4404–4417. [CrossRef]
- Santos Carvalho, R.; Scapulatempo-Neto, C.; Curado, M.P.; de Castro Capuzzo, R.; Marsico Teixeira, F.; Cardoso Pires, R.; Cirino, M.T.; CambreaJoaquim Martins, J.; Almeida Oliveira da Silva, I.; Oliveira, M.A.; et al. HPV-Induced Oropharyngeal Squamous Cell Carcinomas in Brazil:Prevalence, Trend, Clinical, and Epidemiologic Characterization. Cancer Epidemiol. Biomark. Prev. 2021, 30, 1697–1707. [CrossRef] [PubMed]
- Benzerdjeb, N.; Tantot, J.; Blanchet, C.; Philouze, P.; Mekki, Y.; Lopez, J.; Devouassoux-Shisheboran, M. Oropharyngeal Squamous CellCarcinoma: P16/P53 Immunohistochemistry as a Strong Predictor of HPV Tumour Status. Histopathology 2021, 79, 381–390. [CrossRef] [PubMed]
- Korsten, L.H.A.; Jansen, F.; Lissenberg-Witte, B.I.; Vergeer, M.; Brakenhoff, R.H.; Leemans, C.R.; Verdonck-de Leeuw, I.M. The Course of Health-Related Quality of Life from Diagnosis to Two Years Follow-up in Patients with Oropharyngeal Cancer: Does HPV Status Matter? Support. CareCancer 2021, 29, 4473–4483. [CrossRef] [PubMed]
- Skovvang, A.; Jensen, J.S.; Garset-Zamani, M.; Carlander, A.; Grønhøj, C.; von Buchwald, C. The Impact of HPV Genotypes on Survival in HPV-Positive Oropharyngeal Squamous Cell Carcinomas: A Systematic Review. Acta Oto-Laryngol. 2021, 141, 724–728. [CrossRef] [PubMed]
- Lu, X.J.D.; Liu, K.Y.P.; Prisman, E.; Wu, J.; Zhu, Y.S.; Poh, C. Prognostic Value and Cost Benefit of HPV Testing for Oropharyngeal CancerPatients. Oral Dis. 2021, in press. [CrossRef]
- Suresh, K.; Shah, P.V.; Coates, S.; Alexiev, B.A.; Samant, S. In Situ Hybridization for High Risk HPV E6/E7 MRNA in Oropharyn- geal SquamousCell Carcinoma. Am. J. Otolaryngol. 2021, 42, 102782. [CrossRef] [PubMed]
- Zito Marino, F.; Ronchi, A.; Stilo, M.; Cozzolino, I.; La Mantia, E.; Colacurci, N.; Colella, G.; Franco, R. Multiplex HPV RNA in Situ Hybridization/P16 Immunohistochemistry: A Novel Approach to Detect Papillomavirus in HPV-Related Cancers. A Novel Multiplex ISH/IHC Assayto Detect HPV. Infect. Agents Cancer 2020, 15, 46. [CrossRef]
- Randén-Brady, R.; Carpén, T.; Jouhi, L.; Syrjänen, S.; Haglund, C.; Tarkkanen, J.; Remes, S.; Mäkitie, A.; Mattila, P.S.; Silén, S.; et al. In Situ Hybridization for High-Risk HPV E6/E7 MRNA Is a Superior Method for Detecting Transcriptionally Active HPV in Oropharyngeal Cancer. Hum.Pathol. 2019, 90, 97–105. [CrossRef]
- Shinn, J.R.; Davis, S.J.; Lang-Kuhs, K.A.; Rohde, S.; Wang, X.; Liu, P.; Dupont, W.D.; Plummer, D.; Thorstad, W.L.; Chernock, R.D.; et al.Oropharyngeal Squamous Cell Carcinoma With Discordant P16 and HPV MRNA Results: Incidence and Characterization in a Large, ContemporaryUnited States Cohort. Am. J. Surg. Pathol. 2021, 45, 951–961. [CrossRef] [PubMed]
- Economopoulou, P.; Koutsodontis, G.; Avgeris, M.; Strati, A.; Kroupis, C.; Pateras, I.; Kirodimos, E.; Giotakis, E.; Kotsantis, I.; Maragoudakis, P.; et al. HPV16 E6/E7 Expression in Circulating Tumor Cells in Oropharyngeal Squamous Cell Cancers: A Pilot Study. PLoS ONE 2019, 14, e0215984.[CrossRef]
- Nemescu, D.; Ursu, R.G.; Nemescu, E.R.; Negura, L. Heterogeneous Distribution of Fetal Microchimerism in Local Breast Cancer Environment. PLoSONE 2016, 11, e0147675. [CrossRef]
- Craig, S.G.; Anderson, L.A.; Moran, M.; Graham, L.; Currie, K.; Rooney, K.; Robinson, M.; Bingham, V.; Cuschieri, K.S.; McQuaid, S.; et al. Comparison of Molecular Assays for HPV Testing in Oropharyngeal Squamous Cell Carcinomas: A Population-Based Study in Northern Ireland.Cancer Epidemiol. Biomark. Prev. 2020, 29, 31–38. [CrossRef] [PubMed]
- Simoens, C.; Gorbaslieva, I.; Gheit, T.; Holzinger, D.; Lucas, E.; Ridder, R.; Rehm, S.; Vermeulen, P.; Lammens, M.; Vanderveken, O.M.; et al. HPV DNA Genotyping, HPV E6*I MRNA Detection, and P16INK4a/Ki-67 Staining in Belgian Head and Neck Cancer Patient Specimens, Collected withinthe HPV-AHEAD Study. Cancer Epidemiol. 2021, 72, 101925. [CrossRef] [PubMed]
- Tagliabue, M.; Mena, M.; Maffini, F.; Gheit, T.; Quirós Blasco, B.; Holzinger, D.; Tous, S.; Scelsi, D.; Riva, D.; Grosso, E.; et al. Role of Human Papillomavirus Infection in Head and Neck Cancer in Italy: The HPV-AHEAD Study. Cancers 2020, 12, 3567. [CrossRef]
- Gheit, T.; Anantharaman, D.; Holzinger, D.; Alemany, L.; Tous, S.; Lucas, E.; Prabhu, P.R.; Pawlita, M.; Ridder, R.; Rehm, S.; et al. Role of Mucosal High-Risk Human Papillomavirus Types in Head and Neck Cancers in Central India. Int. J. Cancer 2017, 141, 143–151. [CrossRef]
- Seiwert, T.Y.; Zuo, Z.; Keck, M.K.; Khattri, A.; Pedamallu, C.S.; Stricker, T.; Brown, C.; Pugh, T.J.; Stojanov, P.; Cho, J.; et al. Integrative and Comparative Genomic Analysis of HPV-Positive and HPV-Negative Head and Neck Squamous Cell Carcinomas. Clin. Cancer Res. 2015, 21, 632–641. [CrossRef]
- Koole, K.; Clausen, M.J.A.M.; van Es, R.J.J.; van Kempen, P.M.W.; Melchers, L.J.; Koole, R.; Langendijk, J.A.; van Diest, P.J.; Roodenburg, J.L.N.; Schuuring, E.; et al. FGFR Family Members Protein Expression as Prognostic Markers in Oral Cavity and Oropharyngeal Squamous CellCarcinoma. Mol. Diagn. Ther. 2016, 20, 363–374. [CrossRef]
- Koole, K.; Brunen, D.; van Kempen, P.M.W.; Noorlag, R.; de Bree, R.; Lieftink, C.; van Es, R.J.J.; Bernards, R.; Willems, S.M. FGFR1 Is aPotential Prognostic Biomarker and Therapeutic Target in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2016, 22, 3884–3893.[CrossRef] [PubMed]
- Bersani, C.; Sivars, L.; Haeggblom, L.; DiLorenzo, S.; Mints, M.; Ährlund-Richter, A.; Tertipis, N.; Munck-Wikland, E.; Näsman, A.; Ramqvist, T.;et al. Targeted Sequencing of Tonsillar and Base of Tongue Cancer and Human Papillomavirus Positive Unknown Primary of the Head and Neck Reveals Prognostic Effects of Mutated FGFR3. Oncotarget 2017, 8, 35339–35350. [CrossRef] [PubMed]
- Lim, S.M.; Cho, S.H.; Hwang, I.G.; Choi, J.W.; Chang, H.; Ahn, M.-J.; Park, K.U.; Kim, J.-W.; Ko, Y.H.; Ahn, H.K.; et al. Investigating the Feasibility of Targeted Next-Generation Sequencing to Guide the Treatment of Head and Neck Squamous Cell Carcinoma. Cancer Res. Treat. 2019,51, 300–312. [CrossRef] [PubMed]
- Sablin, M.-P.; Dubot, C.; Klijanienko, J.; Vacher, S.; Ouafi, L.; Chemlali, W.; Caly, M.; Sastre-Garau, X.; Lappartient, E.; Mariani, O.; et al.Identification of New Candidate Therapeutic Target Genes in Head and Neck Squamous Cell Carcinomas. Oncotarget 2016, 7, 47418–47430. [CrossRef]
- Feldman, R.; Gatalica, Z.; Knezetic, J.; Reddy, S.; Nathan, C.-A.; Javadi, N.; Teknos, T. Molecular Profiling of Head and Neck Squamous CellCarcinoma. Head Neck 2016, 38 (Suppl. 1), E1625–E1638. [CrossRef] [PubMed]
- Gillison, M.L.; Akagi, K.; Xiao, W.; Jiang, B.; Pickard, R.K.L.; Li, J.; Swanson, B.J.; Agrawal, A.D.; Zucker, M.; Stache-Crain, B.; et al. Human Papillomavirus and the Landscape of Secondary Genetic Alterations in Oral Cancers. Genome Res. 2019, 29, 1–17. [CrossRef] [PubMed]
- Farah, C.S. Molecular Landscape of Head and Neck Cancer and Implications for Therapy. Ann. Transl. Med. 2021, 9, 915. [CrossRef]
- Holzhauser, S.; Lukoseviciute, M.; Andonova, T.; Ursu, R.G.; Dalianis, T.; Wickström, M.; Kostopoulou, O.N. Targeting Fibroblast Growth Factor Receptor (FGFR) and Phosphoinositide 3-Kinase (PI3K) Signaling Pathways in Medulloblastoma Cell Lines. Anticancer Res. 2020, 40, 53–66.[CrossRef]
- Holzhauser, S.; Wild, N.; Zupancic, M.; Ursu, R.G.; Bersani, C.; Näsman, A.; Kostopoulou, O.N.; Dalianis, T. Targeted Therapy With PI3K and FGFR Inhibitors on Human Papillomavirus Positive and Negative Tonsillar and Base of Tongue Cancer Lines With and Without CorrespondingMutations. Front. Oncol. 2021, 11, 640490. [CrossRef] [PubMed]
- Ganci, F.; Pulito, C.; Valsoni, S.; Sacconi, A.; Turco, C.; Vahabi, M.; Manciocco, V.; Mazza, E.M.C.; Meens, J.; Karamboulas, C.; et al. PI3KInhibitors Curtail MYC-Dependent Mutant P53 Gain-of-Function in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2020, 26, 2956–2971. [CrossRef]
- Beaty, B.T.; Moon, D.H.; Shen, C.J.; Amdur, R.J.; Weiss, J.; Grilley-Olson, J.; Patel, S.; Zanation, A.; Hackman, T.G.; Thorp, B.; et al. PIK3CA Mutation in HPV-Associated OPSCC Patients Receiving Deintensified Chemoradiation. J. Natl. Cancer Inst. 2020, 112, 855–858. [CrossRef]
- Geiger, J.L.; Bauman, J.E.; Gibson, M.K.; Gooding, W.E.; Varadarajan, P.; Kotsakis, A.; Martin, D.; Gutkind, J.S.; Hedberg, M.L.; Grandis, J.R.; et al. Phase II Trial of Everolimus in Patients with Previously Treated Recurrent or Metastatic Head and Neck Squamous Cell Carcinoma. Head Neck 2016, 38, 1759–1764. [CrossRef] [PubMed]
- Lewis, J.S. Human Papillomavirus Testing in Head and Neck Squamous Cell Carcinoma in 2020: Where Are We Now and Where Are We Going?Head Neck Pathol. 2020, 14, 321–329. [CrossRef] [PubMed]
- Näsman, A.; Du, J.; Dalianis, T. A Global Epidemic Increase of an HPV-Induced Tonsil and Tongue Base Cancer—Potential Benefit from a Pan-Gender Use of HPV Vaccine. J. Intern. Med. 2020, 287, 134–152. [CrossRef] [PubMed]
- Stern, P.L.; Dalianis, T. Oropharyngeal Squamous Cell Carcinoma Treatment in the Era of Immune Checkpoint Inhibitors. Viruses 2021, 13, 1234. [CrossRef]